Virtual karyotype: Difference between revisions
No edit summary |
No edit summary |
||
| Line 5: | Line 5: | ||
==What is a Virtual Karyotype?== |
==What is a Virtual Karyotype?== |
||
Recently, platforms for generating high-resolution karyotypes ''in silico'' from disrupted DNA have emerged, such as [[array comparative genomic hybridization]] (arrayCGH) and [[SNP array]]s. Conceptually, the arrays are composed of hundreds to millions of probes which are complementary to a region of interest in the genome. The disrupted DNA from the test sample is fragmented, labeled, and hybridized to the array. The hybridization signal intensities for each probe are used specialize software to generate a log2ratio of test/normal for each probe on the array. Knowing the address of each probe on the array and the address of each probe in the genome, the software lines the probes up in chromosomal order and reconstructs the genome ''in silico''. |
Recently, platforms for generating high-resolution karyotypes ''in silico'' from disrupted DNA have emerged, such as [[array comparative genomic hybridization]] (arrayCGH) and [[SNP array]]s. Conceptually, the arrays are composed of hundreds to millions of probes which are complementary to a region of interest in the genome. The disrupted DNA from the test sample is fragmented, labeled, and hybridized to the array. The hybridization signal intensities for each probe are used specialize software to generate a log2ratio of test/normal for each probe on the array. Knowing the address of each probe on the array and the address of each probe in the genome, the software lines the probes up in chromosomal order and reconstructs the genome ''in silico''. |
||
Virtual karyptypes can be performed on germline samples for constitutional disorders, and clinical testing is available from dozens of different CLIA certified laboratories ([http://www.genetests.org/query?testid=234754 GeneTests.org] |
|||
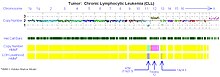
[[Image:CLL_ForWiki.jpg|thumb|Virtual karyotype of chronic lymphocytic leukemia using SNP array.]] |
[[Image:CLL_ForWiki.jpg|thumb|Virtual karyotype of chronic lymphocytic leukemia using SNP array.]] |
||
Array-based karyotyping can be done with several different platforms, both laboratory-developed and commercial. The arrays themselves can be genome-wide (probes distributed over the entire genome) or targeted (probes for genomic regions known to be involved in a specific disease) or a combination of both. Further, arrays used for karyotyping may use non-polymorphic probes, polymorphic probes (i.e., SNP-containing), or a combination of both. Non-polymorphic probes can provide only copy number information, while SNP arrays can provide both copy number and loss-of-heterozygosity (LOH) status in one assay. The probe types used for non-polymorphic arrays include cDNA, BAC clones (e.g., [http://www.cytochip.com BlueGnome]), and oligonucleotides (e.g., [[Agilent]], Santa Clara, CA, USA or [[Nimblegen]], Madison, WI, USA). Commercially available oligonucleotide SNP arrays can be solid phase ([[Affymetrix]], Santa Clara, CA, USA) or bead-based ([[Illumina]], SanDiego, CA, USA). Despite the diversity of platforms, ultimately they all use genomic DNA from disrupted cells to recreate a high resolution karyotype ''in silico''. The end product does not yet have a consistent name, and has been called virtual karyotyping<ref>Hagenkord JM, Parwani AV, Lyons-Weiler MA, Alvarez K, Amato R, Gatalica Z, et al. Virtual karyotyping with SNP microarrays reduces uncertainty in the diagnosis of renal epithelial tumors. Diagnostic pathology. 2008;3:44. [Epub ahead of print]</ref><ref>Monzon FA, Hagenkord JM, Lyons-Weiler MA, Balani JP, Parwani AV, Sciulli CM, et al. Whole genome SNP arrays as a potential diagnostic tool for the detection of characteristic chromosomal aberrations in renal epithelial tumors. Mod Pathol. 2008 May;21(5):599-608.</ref> digital karyotyping<ref>Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008 October 21, 2008;105(42):16224-9.</ref>, molecular allelokaryotyping<ref>Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, et al. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer. 2008 Mar 15;112(6):1296-305.</ref>, and molecular karyotyping<ref>Vermeesch JR, Fiegler H, de Leeuw N, Szuhai K, Schoumans J, Ciccone R, et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur J Hum Genet. 2007;15(11):1105-14.</ref>. Other terms used to describe the arrays used for karyotyping include SOMA (SNP oligonucleotide microarrays)<ref>Kulharya AS, Flannery DB, Norris K, Lovell C, Levy B, Velagaleti GV. Fine mapping of breakpoints in two unrelated patients with rare overlapping interstitial deletions of 9q with mild dysmorphic features. American journal of medical genetics. 2008 Sep 1;146A(17):2234-41.</ref> and CMA (chromosome microarray)<ref>Nowakowska B, Stankiewicz P, Obersztyn E, Ou Z, Li J, Chinault AC, et al. Application of metaphase HR-CGH and targeted Chromosomal Microarray Analyses to genomic characterization of 116 patients with mental retardation and dysmorphic features. American journal of medical genetics. 2008 Sep 15;146A(18):2361-9.</ref><ref>Probst FJ, Roeder ER, Enciso VB, Ou Z, Cooper ML, Eng P, et al. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. American journal of medical genetics. 2007 Jun 15;143A(12):1358-65.</ref>. Some consider all platforms to be a type of [[array comparative genomic hybridization]] (arrayCGH), while others reserve that term for two-dye methods, and still others who segregate SNP arrays because they generate more and different information than two-dye arrayCGH methods. |
Array-based karyotyping can be done with several different platforms, both laboratory-developed and commercial. The arrays themselves can be genome-wide (probes distributed over the entire genome) or targeted (probes for genomic regions known to be involved in a specific disease) or a combination of both. Further, arrays used for karyotyping may use non-polymorphic probes, polymorphic probes (i.e., SNP-containing), or a combination of both. Non-polymorphic probes can provide only copy number information, while SNP arrays can provide both copy number and loss-of-heterozygosity (LOH) status in one assay. The probe types used for non-polymorphic arrays include cDNA, BAC clones (e.g., [http://www.cytochip.com BlueGnome]), and oligonucleotides (e.g., [[Agilent]], Santa Clara, CA, USA or [[Nimblegen]], Madison, WI, USA). Commercially available oligonucleotide SNP arrays can be solid phase ([[Affymetrix]], Santa Clara, CA, USA) or bead-based ([[Illumina]], SanDiego, CA, USA). Despite the diversity of platforms, ultimately they all use genomic DNA from disrupted cells to recreate a high resolution karyotype ''in silico''. The end product does not yet have a consistent name, and has been called virtual karyotyping<ref>Hagenkord JM, Parwani AV, Lyons-Weiler MA, Alvarez K, Amato R, Gatalica Z, et al. Virtual karyotyping with SNP microarrays reduces uncertainty in the diagnosis of renal epithelial tumors. Diagnostic pathology. 2008;3:44. [Epub ahead of print]</ref><ref>Monzon FA, Hagenkord JM, Lyons-Weiler MA, Balani JP, Parwani AV, Sciulli CM, et al. Whole genome SNP arrays as a potential diagnostic tool for the detection of characteristic chromosomal aberrations in renal epithelial tumors. Mod Pathol. 2008 May;21(5):599-608.</ref> digital karyotyping<ref>Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008 October 21, 2008;105(42):16224-9.</ref>, molecular allelokaryotyping<ref>Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, et al. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer. 2008 Mar 15;112(6):1296-305.</ref>, and molecular karyotyping<ref>Vermeesch JR, Fiegler H, de Leeuw N, Szuhai K, Schoumans J, Ciccone R, et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur J Hum Genet. 2007;15(11):1105-14.</ref>. Other terms used to describe the arrays used for karyotyping include SOMA (SNP oligonucleotide microarrays)<ref>Kulharya AS, Flannery DB, Norris K, Lovell C, Levy B, Velagaleti GV. Fine mapping of breakpoints in two unrelated patients with rare overlapping interstitial deletions of 9q with mild dysmorphic features. American journal of medical genetics. 2008 Sep 1;146A(17):2234-41.</ref> and CMA (chromosome microarray)<ref>Nowakowska B, Stankiewicz P, Obersztyn E, Ou Z, Li J, Chinault AC, et al. Application of metaphase HR-CGH and targeted Chromosomal Microarray Analyses to genomic characterization of 116 patients with mental retardation and dysmorphic features. American journal of medical genetics. 2008 Sep 15;146A(18):2361-9.</ref><ref>Probst FJ, Roeder ER, Enciso VB, Ou Z, Cooper ML, Eng P, et al. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. American journal of medical genetics. 2007 Jun 15;143A(12):1358-65.</ref>. Some consider all platforms to be a type of [[array comparative genomic hybridization]] (arrayCGH), while others reserve that term for two-dye methods, and still others who segregate SNP arrays because they generate more and different information than two-dye arrayCGH methods. |
||
==What Types of Genetic Aberrations Can Be Detected by Virtual Karyotypes?== |
|||
Revision as of 19:04, 9 March 2009
What is a Karyotype?
A karyotype is the characteristic chromosome complement of a eukaryote species.[1][2] A karyotype is typically presented as an image of the chromosomes from a single cell arranged from largest (chromosome 1) to smallest (chromosome 22), with the sex chromosomes (X and/or Y) shown last. Historically, karyotypes have been obtained by staining cells after they have been chemically arrested during cell division. Karyotypes have been used for several decades to identify chromosomal abnormalities in both germline and cancer cells. Conventional karyotypes can assess the entire genome for changes in chromosome structure and number, but the resolution is relatively coarse with a detection limit of >3Mb.[3]

What is a Virtual Karyotype?
Recently, platforms for generating high-resolution karyotypes in silico from disrupted DNA have emerged, such as array comparative genomic hybridization (arrayCGH) and SNP arrays. Conceptually, the arrays are composed of hundreds to millions of probes which are complementary to a region of interest in the genome. The disrupted DNA from the test sample is fragmented, labeled, and hybridized to the array. The hybridization signal intensities for each probe are used specialize software to generate a log2ratio of test/normal for each probe on the array. Knowing the address of each probe on the array and the address of each probe in the genome, the software lines the probes up in chromosomal order and reconstructs the genome in silico.
Virtual karyptypes can be performed on germline samples for constitutional disorders, and clinical testing is available from dozens of different CLIA certified laboratories (GeneTests.org
Array-based karyotyping can be done with several different platforms, both laboratory-developed and commercial. The arrays themselves can be genome-wide (probes distributed over the entire genome) or targeted (probes for genomic regions known to be involved in a specific disease) or a combination of both. Further, arrays used for karyotyping may use non-polymorphic probes, polymorphic probes (i.e., SNP-containing), or a combination of both. Non-polymorphic probes can provide only copy number information, while SNP arrays can provide both copy number and loss-of-heterozygosity (LOH) status in one assay. The probe types used for non-polymorphic arrays include cDNA, BAC clones (e.g., BlueGnome), and oligonucleotides (e.g., Agilent, Santa Clara, CA, USA or Nimblegen, Madison, WI, USA). Commercially available oligonucleotide SNP arrays can be solid phase (Affymetrix, Santa Clara, CA, USA) or bead-based (Illumina, SanDiego, CA, USA). Despite the diversity of platforms, ultimately they all use genomic DNA from disrupted cells to recreate a high resolution karyotype in silico. The end product does not yet have a consistent name, and has been called virtual karyotyping[4][5] digital karyotyping[6], molecular allelokaryotyping[7], and molecular karyotyping[8]. Other terms used to describe the arrays used for karyotyping include SOMA (SNP oligonucleotide microarrays)[9] and CMA (chromosome microarray)[10][11]. Some consider all platforms to be a type of array comparative genomic hybridization (arrayCGH), while others reserve that term for two-dye methods, and still others who segregate SNP arrays because they generate more and different information than two-dye arrayCGH methods.
What Types of Genetic Aberrations Can Be Detected by Virtual Karyotypes?
- ^ White M.J.D. 1973. The chromosomes. 6th ed, Chapman & Hall, London. p28
- ^ Stebbins G.L. 1950. Variation and evolution in plants. Chapter XII: The Karyotype. Columbia University Press N.Y.
- ^ Lars Feuk ARC, Stephen W. Scherer. Structural variation in the human genome. Nature Reviews Genetics. February 2006 2006;7:85-97.
- ^ Hagenkord JM, Parwani AV, Lyons-Weiler MA, Alvarez K, Amato R, Gatalica Z, et al. Virtual karyotyping with SNP microarrays reduces uncertainty in the diagnosis of renal epithelial tumors. Diagnostic pathology. 2008;3:44. [Epub ahead of print]
- ^ Monzon FA, Hagenkord JM, Lyons-Weiler MA, Balani JP, Parwani AV, Sciulli CM, et al. Whole genome SNP arrays as a potential diagnostic tool for the detection of characteristic chromosomal aberrations in renal epithelial tumors. Mod Pathol. 2008 May;21(5):599-608.
- ^ Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008 October 21, 2008;105(42):16224-9.
- ^ Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, et al. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer. 2008 Mar 15;112(6):1296-305.
- ^ Vermeesch JR, Fiegler H, de Leeuw N, Szuhai K, Schoumans J, Ciccone R, et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur J Hum Genet. 2007;15(11):1105-14.
- ^ Kulharya AS, Flannery DB, Norris K, Lovell C, Levy B, Velagaleti GV. Fine mapping of breakpoints in two unrelated patients with rare overlapping interstitial deletions of 9q with mild dysmorphic features. American journal of medical genetics. 2008 Sep 1;146A(17):2234-41.
- ^ Nowakowska B, Stankiewicz P, Obersztyn E, Ou Z, Li J, Chinault AC, et al. Application of metaphase HR-CGH and targeted Chromosomal Microarray Analyses to genomic characterization of 116 patients with mental retardation and dysmorphic features. American journal of medical genetics. 2008 Sep 15;146A(18):2361-9.
- ^ Probst FJ, Roeder ER, Enciso VB, Ou Z, Cooper ML, Eng P, et al. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. American journal of medical genetics. 2007 Jun 15;143A(12):1358-65.
