Kinesin
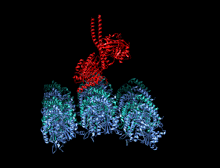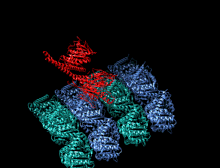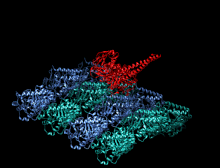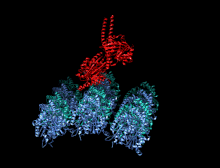

A kinesin is a protein belonging to a class of motor proteins found in eukaryotic cells. Kinesins move along microtubule (MT) filaments and are powered by the hydrolysis of adenosine triphosphate (ATP) (thus kinesins are ATPases, a type of enzyme). The active movement of kinesins supports several cellular functions including mitosis, meiosis and transport of cellular cargo, such as in axonal transport, and intraflagellar transport. Most kinesins walk towards the plus end of a microtubule, which, in most cells, entails transporting cargo such as protein and membrane components from the center of the cell towards the periphery.[1] This form of transport is known as anterograde transport. In contrast, dyneins are motor proteins that move toward the minus end of a microtubule in retrograde transport.
Discovery
The first kinesins to be discovered were microtubule-based anterograde intracellular transport motors[2] in 1985, based on their motility in cytoplasm extruded from the giant axon of the squid.[3]
The founding member of this superfamily, kinesin-1, was isolated as a heterotetrameric fast axonal organelle transport motor consisting of four parts: two identical motor subunits (called Kinesin Heavy Chain (KHC) molecules) and two other molecules each known as a Kinesin Light Chain (KLC). These were discovered via microtubule affinity purification from neuronal cell extracts.[4] Subsequently, a different, heterotrimeric plus-end-directed MT-based motor named kinesin-2, consisting of two distinct KHC-related motor subunits and an accessory "KAP" subunit, was purified from echinoderm egg/embryo extracts[5] and is best known for its role in transporting protein complexes (intraflagellar transport particles) along axonemes during ciliogenesis.[6] Molecular genetic and genomic approaches have led to the recognition that the kinesins form a diverse superfamily of motors that are responsible for multiple intracellular motility events in eukaryotic cells.[7][8][9][10] For example, the genomes of mammals encode more than 40 kinesin proteins,[11] organized into at least 14 families named kinesin-1 through kinesin-14.[12]
Structure
Overall structure
Members of the kinesin superfamily vary in shape but the prototypical kinesin-1 motor consists of two Kinesin Heavy Chain (KHC) molecules which form a protein dimer (molecule pair) that binds two light chains (KLCs), which are unique for different cargos.
The heavy chain of kinesin-1 comprises a globular head (the motor domain) at the amino terminal end connected via a short, flexible neck linker to the stalk – a long, central alpha-helical coiled coil domain – that ends in a carboxy terminal tail domain which associates with the light-chains. The stalks of two KHCs intertwine to form a coiled coil that directs dimerization of the two KHCs. In most cases transported cargo binds to the kinesin light chains, at the TPR motif sequence of the KLC, but in some cases cargo binds to the C-terminal domains of the heavy chains.[13]
Kinesin motor domain
| Kinesin motor domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
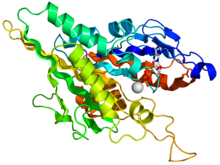 Crystallographic structure of the human kinesin motor domain depicted as a rainbow colored cartoon (N-terminus = blue, C-terminus = red) complexed with ADP (stick diagram, carbon = white, oxygen = red, nitrogen = blue, phosphorus = orange) and a magnesium ion (grey sphere).[14] | |||||||||
| Identifiers | |||||||||
| Symbol | Kinesin motor domain | ||||||||
| Pfam | PF00225 | ||||||||
| InterPro | IPR001752 | ||||||||
| SMART | SM00129 | ||||||||
| PROSITE | PS50067 | ||||||||
| SCOP2 | 1bg2 / SCOPe / SUPFAM | ||||||||
| CDD | cd00106 | ||||||||
| |||||||||
The head is the signature of kinesin and its amino acid sequence is well conserved among various kinesins. Each head has two separate binding sites: one for the microtubule and the other for ATP. ATP binding and hydrolysis as well as ADP release change the conformation of the microtubule-binding domains and the orientation of the neck linker with respect to the head; this results in the motion of the kinesin. Several structural elements in the Head, including a central beta-sheet domain and the Switch I and II domains, have been implicated as mediating the interactions between the two binding sites and the neck domain. Kinesins are structurally related to G proteins, which hydrolyze GTP instead of ATP. Several structural elements are shared between the two families, notably the Switch I and Switch II domain.

Basic kinesin regulation
Kinesins tend to have low basal enzymatic activity which becomes significant when microtubule-activated.[16] In addition, many members of the kinesin superfamily can be self-inhibited by the binding of tail domain to the motor domain.[17] Such self-inhibition can then be relieved via additional regulation such as binding to cargo, cargo adapters or other microtubule-associated proteins.[18][19][20]
Cargo transport
In the cell, small molecules, such as gases and glucose, diffuse to where they are needed. Large molecules synthesised in the cell body, intracellular components such as vesicles and organelles such as mitochondria are too large (and the cytosol too crowded) to be able to diffuse to their destinations. Motor proteins fulfill the role of transporting large cargo about the cell to their required destinations. Kinesins are motor proteins that transport such cargo by walking unidirectionally along microtubule tracks hydrolysing one molecule of adenosine triphosphate (ATP) at each step.[21] It was thought that ATP hydrolysis powered each step, the energy released propelling the head forwards to the next binding site.[22] However, it has been proposed that the head diffuses forward and the force of binding to the microtubule is what pulls the cargo along.[23] In addition viruses, HIV for example, exploit kinesins to allow virus particle shuttling after assembly.[24]
There is significant evidence that cargoes in-vivo are transported by multiple motors.[25][26][27][28]
Direction of motion
Motor proteins travel in a specific direction along a microtubule. Microtubules are polar; meaning, the heads only bind to the microtubule in one orientation, while ATP binding gives each step its direction through a process known as neck linker zippering.[29]
It has been previously known that kinesin move cargo towards the plus (+) end of a microtubule, also known as anterograde transport/orthograde transport.[30] However, it has been recently discovered that in budding yeast cells kinesin Cin8 (a member of the Kinesin-5 family) can move toward the minus end as well, or retrograde transport. This means, these unique yeast kinesin homotetramers have the novel ability to move bi-directionally.[31][32][33] Kinesin, so far, has only been shown to move toward the minus end when in a group, with motors sliding in the antiparallel direction in an attempt to separate microtubules.[34] This dual directionality has been observed in identical conditions where free Cin8 molecules move towards the minus end, but cross-linking Cin8 move toward the plus ends of each cross-linked microtubule. One specific study tested the speed at which Cin8 motors moved, their results yielded a range of about 25-55 nm/s, in the direction of the spindle poles.[35] On an individual basis it has been found that by varying ionic conditions Cin8 motors can become as fast as 380 nm/s.[35] It is suggested that the bidirectionality of yeast kinesin-5 motors such as Cin8 and Cut7 is a result of coupling with other Cin8 motors and helps to fulfill the role of dynein in budding yeast, as opposed to the human homologue of these motors, the plus directed Eg5.[36] This discovery in kinesin-14 family proteins (such as Drosophila melanogaster NCD, budding yeast KAR3, and Arabidopsis thaliana ATK5) allows kinesin to walk in the opposite direction, toward microtubule minus end.[37] This is not typical of kinesin, rather, an exception to the normal direction of movement.

Another type of motor protein, known as dyneins, move towards the minus end of the microtubule. Thus, they transport cargo from the periphery of the cell towards the center. An example of this would be transport occurring from the terminal boutons of a neuronal axon to the cell body (soma). This is known as retrograde transport.
Mechanism of movement
In 2023 direct visualization of kinesin "walking" along a microtubule, in real-time was reported.[38] In a "hand-over-hand" mechanism, the kinesin heads step past one another, alternating the lead position. Thus in each step the leading head becomes the trailing head, while the trailing head becomes the leading head.
- This cycle begins with the trailing head releasing inorganic phosphate (Pi) derived from the hydrolysis of ATP.
- The trailing head detaches from the microtubule and rotates into its rightward displaced unbound state.
- The leading head binds ATP which causes the neck linker to dock to it, which moves the trailing head around the leading head into a position further along the microtubule in the direction of travel. The trailing head remains unbound.
- The ATP in the leading head is hydrolyzed.
- The trailing head releases its ADP and the binds to the microtubule becoming the leading head.[39][40][41][42][43][44]
Theoretical modeling
A number of theoretical models of the molecular motor protein kinesin have been proposed.[45][46][47] Many challenges are encountered in theoretical investigations given the remaining uncertainties about the roles of protein structures, the precise way energy from ATP is transformed into mechanical work, and the roles played by thermal fluctuations. This is a rather active area of research. There is a need especially for approaches which better make a link with the molecular architecture of the protein and data obtained from experimental investigations.
The single-molecule dynamics are already well described[48] but it seems that these nano scale machines typically work in large teams.
Single-molecule dynamics are based on the distinct chemical states of the motor and observations about its mechanical steps.[49] For small concentrations of adenosine diphosphate, the motor's behaviour is governed by the competition of two chemomechanical motor cycles which determine the motor's stall force. A third cycle becomes important for large ADP concentrations.[49] Models with a single cycle have been discussed too. Seiferth et al. demonstrated how quantities such as the velocity or the entropy production of a motor change when adjacent states are merged in a multi-cyclic model until eventually the number of cycles is reduced.[50]
Recent experimental research has shown that kinesins, while moving along microtubules, interact with each other,[51][52] the interactions being short range and weak attractive (1.6±0.5 KBT). One model that has been developed takes into account these particle interactions,[48] where the dynamic rates change accordingly with the energy of interaction. If the energy is positive the rate of creating bonds (q) will be higher while the rate of breaking bonds (r) will be lower. One can understand that the rates of entrance and exit in the microtubule will be changed as well by the energy (See figure 1 in reference 30). If the second site is occupied the rate of entrance will be α*q and if the last but one site is occupied the rate of exit will be β*r. This theoretical approach agrees with the results of Monte Carlo simulations for this model, especially for the limiting case of very large negative energy. The normal totally asymmetric simple exclusion process for (or TASEP) results can be recovered from this model making the energy equal to zero.
Mitosis
In recent years, it has been found that microtubule-based molecular motors (including a number of kinesins) have a role in mitosis (cell division). Kinesins are important for proper spindle length and are involved in sliding microtubules apart within the spindle during prometaphase and metaphase, as well as depolymerizing microtubule minus ends at centrosomes during anaphase.[53] Specifically, Kinesin-5 family proteins act within the spindle to slide microtubules apart, while the Kinesin 13 family act to depolymerize microtubules.
Kinesin superfamily
Human kinesin superfamily members include the following proteins, which in the standardized nomenclature developed by the community of kinesin researchers, are organized into 14 families named kinesin-1 through kinesin-14:[12]
- 1A – KIF1A, 1B – KIF1B, 1C – KIF1C = kinesin-3
- 2A – KIF2A, 2C – KIF2C = kinesin-13
- 3B – KIF3B or 3C – KIF3C ,3A - KIF3A = kinesin-2
- 4A – KIF4A, 4B – KIF4B = kinesin-4
- 5A – KIF5A, 5B – KIF5B, 5C – KIF5C = kinesin-1
- 6 – KIF6 = kinesin-9
- 7 – KIF7 = kinesin-4
- 9 – KIF9 = kinesin-9
- 11 – KIF11 = kinesin-5
- 12 – KIF12 = kinesin-12
- 13A – KIF13A, 13B – KIF13B = kinesin-3
- 14 – KIF14 = kinesin-3
- 15 – KIF15 = kinesin-12
- 16B – KIF16B = kinesin-3
- 17 – KIF17 = kinesin-2
- 18A – KIF18A, 18B – KIF18B = kinesin-8
- 19 – KIF19 = kinesin-8
- 20A – KIF20A, 20B – KIF20B = kinesin-6
- 21A – KIF21A, 21B – KIF21B = kinesin-4
- 22 – KIF22 = kinesin-10
- 23 – KIF23 = kinesin-6
- 24 – KIF24 = kinesin-13
- 25 – KIF25 = kinesin-14
- 26A – KIF26A, 26B – KIF26B = kinesin-11
- 27 – KIF27 = kinesin-4
- C1 – KIFC1, C2 – KIFC2, C3 – KIFC3 = kinesin-14
kinesin-1 light chains:
kinesin-2 associated protein:
- KIFAP3 (also known as KAP-1, KAP3)
See also
- Axonal transport
- Dynein
- Intraflagellar transport along cilia
- Kinesin 8
- Kinesin 13
- KRP
- Molecular motor
- Transport by multiple-motor proteins
References
- ^ Berg J, Tymoczko JL, Stryer L (2002). "Kinesin and Dynein Move Along Microtubules". Biochemistry. 5th Edition.
- ^ Vale RD (February 2003). "The molecular motor toolbox for intracellular transport". Cell. 112 (4): 467–80. doi:10.1016/S0092-8674(03)00111-9. PMID 12600311. S2CID 15100327.
- ^ Endow SA, Kull FJ, Liu H (October 2010). "Kinesins at a glance". Journal of Cell Science. 123 (Pt 20): 3420–4. doi:10.1242/jcs.064113. PMC 2951464. PMID 20930137.
- ^ Vale RD, Reese TS, Sheetz MP (August 1985). "Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility". Cell. 42 (1): 39–50. doi:10.1016/S0092-8674(85)80099-4. PMC 2851632. PMID 3926325.
- ^ Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM (November 1993). "Novel heterotrimeric kinesin-related protein purified from sea urchin eggs". Nature. 366 (6452): 268–70. Bibcode:1993Natur.366..268C. doi:10.1038/366268a0. PMID 8232586. S2CID 4367715.
- ^ Rosenbaum JL, Witman GB (November 2002). "Intraflagellar transport". Nature Reviews. Molecular Cell Biology. 3 (11): 813–25. doi:10.1038/nrm952. PMID 12415299. S2CID 12130216.
- ^ Yang JT, Laymon RA, Goldstein LS (March 1989). "A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analyses". Cell. 56 (5): 879–89. doi:10.1016/0092-8674(89)90692-2. PMID 2522352. S2CID 44318695.
- ^ Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N (December 1992). "Kinesin family in murine central nervous system". The Journal of Cell Biology. 119 (5): 1287–96. doi:10.1083/jcb.119.5.1287. PMC 2289715. PMID 1447303.
- ^ Enos AP, Morris NR (March 1990). "Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans". Cell. 60 (6): 1019–27. doi:10.1016/0092-8674(90)90350-N. PMID 2138511. S2CID 27420513.
- ^ Meluh PB, Rose MD (March 1990). "KAR3, a kinesin-related gene required for yeast nuclear fusion". Cell. 60 (6): 1029–41. doi:10.1016/0092-8674(90)90351-E. PMID 2138512. S2CID 19660190.
- ^ Hirokawa N, Noda Y, Tanaka Y, Niwa S (October 2009). "Kinesin superfamily motor proteins and intracellular transport". Nature Reviews. Molecular Cell Biology. 10 (10): 682–96. doi:10.1038/nrm2774. PMID 19773780. S2CID 18129292.
- ^ a b Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L (October 2004). "A standardized kinesin nomenclature". The Journal of Cell Biology. 167 (1): 19–22. doi:10.1083/jcb.200408113. PMC 2041940. PMID 15479732.
- ^ Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS (March 1989). "Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration". Cell. 56 (5): 867–78. doi:10.1016/0092-8674(89)90691-0. PMID 2522351. S2CID 731898.
- ^ PDB: 1BG2; Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD (April 1996). "Crystal structure of the kinesin motor domain reveals a structural similarity to myosin". Nature. 380 (6574): 550–5. Bibcode:1996Natur.380..550J. doi:10.1038/380550a0. PMC 2851642. PMID 8606779.
- ^ a b Kaan HY, Hackney DD, Kozielski F (August 2011). "The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition". Science. 333 (6044): 883–5. Bibcode:2011Sci...333..883K. doi:10.1126/science.1204824. PMC 3339660. PMID 21836017.
- ^ Stewart RJ, Thaler JP, Goldstein LS (June 1993). "Direction of microtubule movement is an intrinsic property of the motor domains of kinesin heavy chain and Drosophila ncd protein". Proceedings of the National Academy of Sciences of the United States of America. 90 (11): 5209–13. Bibcode:1993PNAS...90.5209S. doi:10.1073/pnas.90.11.5209. PMC 46685. PMID 8506368.
- ^ Verhey KJ, Hammond JW (November 2009). "Traffic control: regulation of kinesin motors". Nature Reviews. Molecular Cell Biology. 10 (11): 765–77. doi:10.1038/nrm2782. PMID 19851335. S2CID 10713993.
- ^ Siddiqui N, Zwetsloot AJ, Bachmann A, Roth D, Hussain H, Brandt J, et al. (June 2019). "PTPN21 and Hook3 relieve KIF1C autoinhibition and activate intracellular transport". Nature Communications. 10 (1): 2693. Bibcode:2019NatCo..10.2693S. doi:10.1038/s41467-019-10644-9. PMC 6584639. PMID 31217419.
- ^ Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ (January 2007). "Two binding partners cooperate to activate the molecular motor Kinesin-1". The Journal of Cell Biology. 176 (1): 11–7. doi:10.1083/jcb.200605099. PMC 2063617. PMID 17200414.
- ^ Hooikaas PJ, Martin M, Mühlethaler T, Kuijntjes GJ, Peeters CA, Katrukha EA, et al. (April 2019). "MAP7 family proteins regulate kinesin-1 recruitment and activation". The Journal of Cell Biology. 218 (4): 1298–1318. doi:10.1083/jcb.201808065. PMC 6446838. PMID 30770434.
- ^ Schnitzer MJ, Block SM (July 1997). "Kinesin hydrolyses one ATP per 8-nm step". Nature. 388 (6640): 386–90. Bibcode:1997Natur.388..386S. doi:10.1038/41111. PMID 9237757. S2CID 4363000.
- ^ Vale RD, Milligan RA (April 2000). "The way things move: looking under the hood of molecular motor proteins". Science. 288 (5463): 88–95. Bibcode:2000Sci...288...88V. doi:10.1126/science.288.5463.88. PMID 10753125.
- ^ Mather WH, Fox RF (October 2006). "Kinesin's biased stepping mechanism: amplification of neck linker zippering". Biophysical Journal. 91 (7): 2416–26. Bibcode:2006BpJ....91.2416M. doi:10.1529/biophysj.106.087049. PMC 1562392. PMID 16844749.
- ^ Gaudin R, de Alencar BC, Jouve M, Bèrre S, Le Bouder E, Schindler M, Varthaman A, Gobert FX, Benaroch P (October 2012). "Critical role for the kinesin KIF3A in the HIV life cycle in primary human macrophages". The Journal of Cell Biology. 199 (3): 467–79. doi:10.1083/jcb.201201144. PMC 3483138. PMID 23091068.
- ^ Gross SP, Vershinin M, Shubeita GT (June 2007). "Cargo transport: two motors are sometimes better than one". Current Biology. 17 (12): R478–86. doi:10.1016/j.cub.2007.04.025. PMID 17580082. S2CID 8791125.
- ^ Hancock WO (August 2008). "Intracellular transport: kinesins working together". Current Biology. 18 (16): R715–7. doi:10.1016/j.cub.2008.07.068. PMID 18727910. S2CID 7540556.
- ^ Kunwar A, Vershinin M, Xu J, Gross SP (August 2008). "Stepping, strain gating, and an unexpected force-velocity curve for multiple-motor-based transport". Current Biology. 18 (16): 1173–83. doi:10.1016/j.cub.2008.07.027. PMC 3385514. PMID 18701289.
- ^ Klumpp S, Lipowsky R (November 2005). "Cooperative cargo transport by several molecular motors". Proceedings of the National Academy of Sciences of the United States of America. 102 (48): 17284–9. arXiv:q-bio/0512011. Bibcode:2005PNAS..10217284K. doi:10.1073/pnas.0507363102. PMC 1283533. PMID 16287974.
- ^ Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD (December 1999). "A structural change in the kinesin motor protein that drives motility". Nature. 402 (6763): 778–84. Bibcode:1999Natur.402..778R. doi:10.1038/45483. PMID 10617199. S2CID 573909.
- ^ Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000). "Kinesin, Dynein, and Intracellular Transport".
{{cite journal}}: Cite journal requires|journal=(help) - ^ Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T (April 2011). "Directional switching of the kinesin Cin8 through motor coupling". Science. 332 (6025): 94–9. Bibcode:2011Sci...332...94R. doi:10.1126/science.1199945. PMID 21350123. S2CID 90739364.
- ^ Fallesen T, Roostalu J, Duellberg C, Pruessner G, Surrey T (November 2017). "Ensembles of Bidirectional Kinesin Cin8 Produce Additive Forces in Both Directions of Movement". Biophysical Journal. 113 (9): 2055–2067. Bibcode:2017BpJ...113.2055F. doi:10.1016/j.bpj.2017.09.006. PMC 5685778. PMID 29117528.
- ^ Edamatsu M (March 2014). "Bidirectional motility of the fission yeast kinesin-5, Cut7". Biochemical and Biophysical Research Communications. 446 (1): 231–4. doi:10.1016/j.bbrc.2014.02.106. PMID 24589736.
- ^ Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T (April 2011). "Directional switching of the kinesin Cin8 through motor coupling". Science. 332 (6025): 94–9. Bibcode:2011Sci...332...94R. doi:10.1126/science.1199945. PMID 21350123. S2CID 90739364.
- ^ a b Gerson-Gurwitz A, Thiede C, Movshovich N, Fridman V, Podolskaya M, Danieli T, et al. (November 2011). "Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry". The EMBO Journal. 30 (24): 4942–54. doi:10.1038/emboj.2011.403. PMC 3243633. PMID 22101328.
- ^ Valentine MT, Fordyce PM, Block SM (December 2006). "Eg5 steps it up!". Cell Division. 1 (1): 31. doi:10.1186/1747-1028-1-31. PMC 1716758. PMID 17173688.
- ^ Ambrose JC, Li W, Marcus A, Ma H, Cyr R (April 2005). "A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis". Molecular Biology of the Cell. 16 (4): 1584–92. doi:10.1091/mbc.e04-10-0935. PMC 1073643. PMID 15659646.
- ^ Fei, Jinyu; Zhou, Ruobu (10 March 2023). "Watching biomolecules stride in real time". Science. 379 (6636): 986–987. doi:10.1126/science.adg8451. PMC 10318587. PMID 36893224.
- ^ Deguchi, Takahiro (10 March 2023). "Direct observation of motor protein stepping in living cells using MINFLUX". Science. 379 (6636): 1010–1015. doi:10.1126/science.ade2676. PMC 7614483. PMID 36893247.
- ^ Wolff, Jan; Scheiderer, Lukas; Engelhardt, Tobias; Engelhardt, Johann; Matthias, Jessica; Hell, Stefan (10 March 2023). "MINFLUX dissects the unimpeded walking of kinesin-1". Science. 379 (6636): 1004–1010. doi:10.1126/science.ade2650. PMID 36893244. S2CID 251162014.
- ^ Yildiz A, Tomishige M, Vale RD, Selvin PR (January 2004). "Kinesin walks hand-over-hand". Science. 303 (5658): 676–8. Bibcode:2004Sci...303..676Y. doi:10.1126/science.1093753. PMID 14684828. S2CID 30529199.
- ^ Asbury CL (February 2005). "Kinesin: world's tiniest biped". Current Opinion in Cell Biology. 17 (1): 89–97. doi:10.1016/j.ceb.2004.12.002. PMID 15661524.
- ^ Sindelar CV, Downing KH (March 2010). "An atomic-level mechanism for activation of the kinesin molecular motors". Proceedings of the National Academy of Sciences of the United States of America. 107 (9): 4111–6. Bibcode:2010PNAS..107.4111S. doi:10.1073/pnas.0911208107. PMC 2840164. PMID 20160108.
- ^ Lay Summary (18 February 2010). "Life's smallest motor, cargo carrier of the cells, moves like a seesaw". PhysOrg.com. Retrieved 31 May 2013.
- ^ Atzberger PJ, Peskin CS (January 2006). "A Brownian Dynamics model of kinesin in three dimensions incorporating the force-extension profile of the coiled-coil cargo tether". Bulletin of Mathematical Biology. 68 (1): 131–60. arXiv:0910.5753. doi:10.1007/s11538-005-9003-6. PMID 16794924. S2CID 13534734.
- ^ Peskin CS, Oster G (April 1995). "Coordinated hydrolysis explains the mechanical behavior of kinesin". Biophysical Journal. 68 (4 Suppl): 202S–210S, discussion 210S–211S. PMC 1281917. PMID 7787069.
- ^ Mogilner A, Fisher AJ, Baskin RJ (July 2001). "Structural changes in the neck linker of kinesin explain the load dependence of the motor's mechanical cycle". Journal of Theoretical Biology. 211 (2): 143–57. Bibcode:2001JThBi.211..143M. doi:10.1006/jtbi.2001.2336. PMID 11419956.
- ^ a b Celis-Garza D, Teimouri H, Kolomeisky AB (2015). "Correlations and symmetry of interactions influence collective dynamics of molecular motors". Journal of Statistical Mechanics: Theory and Experiment. 2015 (4): P04013. arXiv:1503.00633. Bibcode:2015JSMTE..04..013C. doi:10.1088/1742-5468/2015/04/p04013. S2CID 14002728.
- ^ a b Liepelt, Steffen; Lipowsky, Reinhard (20 June 2007). "Kinesin's Network of Chemomechanical Motor Cycles". Physical Review Letters. 98 (25): 258102. Bibcode:2007PhRvL..98y8102L. doi:10.1103/PhysRevLett.98.258102. PMID 17678059.
- ^ Seiferth, David; Sollich, Peter; Klumpp, Stefan (29 December 2020). "Coarse graining of biochemical systems described by discrete stochastic dynamics". Physical Review E. 102 (6): 062149. arXiv:2102.13394. Bibcode:2020PhRvE.102f2149S. doi:10.1103/PhysRevE.102.062149. PMID 33466014. S2CID 231652939.
- ^ Seitz A, Surrey T (January 2006). "Processive movement of single kinesins on crowded microtubules visualized using quantum dots". The EMBO Journal. 25 (2): 267–77. doi:10.1038/sj.emboj.7600937. PMC 1383520. PMID 16407972.
- ^ Vilfan A, Frey E, Schwabl F, Thormählen M, Song YH, Mandelkow E (October 2001). "Dynamics and cooperativity of microtubule decoration by the motor protein kinesin". Journal of Molecular Biology. 312 (5): 1011–26. doi:10.1006/jmbi.2001.5020. PMID 11580246.
- ^ Goshima G, Vale RD (August 2005). "Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells". Molecular Biology of the Cell. 16 (8): 3896–907. doi:10.1091/mbc.E05-02-0118. PMC 1182325. PMID 15958489.
Further reading
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L (October 2004). "A standardized kinesin nomenclature". The Journal of Cell Biology. 167 (1): 19–22. doi:10.1083/jcb.200408113. PMC 2041940. PMID 15479732.
External links
- MBInfo - Kinesin transports cargo along microtubules
- Animated model of kinesin walking
- Ron Vale's Seminar: "Molecular Motor Proteins"
- Animation of kinesin movement ASCB image library
- Murphy, V.F. (12 May 2004). "Microtubule Based Movement". tissue.medicalengineer.co.uk. Archived from the original on 22 July 2007. Retrieved 10 December 2015.
- The Inner Life of a Cell, 3D animation featuring a Kinesin transporting a vesicle Archived 7 December 2008 at the Wayback Machine
- The Kinesin Homepage
- Kinesin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- EC 3.6.4.4
- EC 3.6.4.5
- 3D electron microscopy structures of kinesin from the EM Data Bank(EMDB)

