Base J
| |
| Names | |
|---|---|
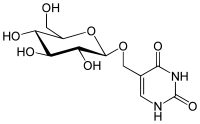
| IUPAC name
5-[[(3R,4S,5S,6R)-3,4,5-Trihydroxy-6-(hydroxymethyl)tetrahydropyran-2-yl]oxymethyl]-1H-pyrimidine-2,4-dione
| |
| Other names
β-D-Glucosyl-5-hydroxymethyluracil
β-D-Glucosyl-hydroxymethyluracil | |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider | |
PubChem CID
|
|
| |
| |
| Properties | |
| C11H16N2O8 | |
| Molar mass | 304.255 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
β-D-Glucopyranosyloxymethyluracil or base J is a hypermodified nucleobase found in the DNA of kinetoplastids including the human pathogenic trypanosomes. It was discovered in 1993, in the trypanosome Trypanosoma brucei and was the first hypermodified nucleobase found in eukaryotic DNA; it has since been found in other kinetoplastids, including Leishmania.[1] Within these organism Base J acts as a RNA polymerase II transcription terminator, with its removal in knockout cells being accompanied by a massive read-through at RNA polymerase II termination sites, which ultimately proves lethal to the cell.[2][3]
Base J is formed by the initial hydroxylation of thymidine and the subsequent glycosylation by an as yet unidentified glycosyltransferase.[1]
References
- ^ a b Borst, Piet; Sabatini, Robert (October 2008). "Base J: Discovery, Biosynthesis, and Possible Functions". Annual Review of Microbiology. 62 (1): 235–251. doi:10.1146/annurev.micro.62.081307.162750. PMID 18729733.
- ^ van Luenen, Henri G.A.M.; Farris, Carol; Jan, Sabrina; Genest, Paul-Andre; Tripathi, Pankaj; Velds, Arno; Kerkhoven, Ron M.; Nieuwland, Marja; Haydock, Andrew; Ramasamy, Gowthaman; Vainio, Saara; Heidebrecht, Tatjana; Perrakis, Anastassis; Pagie, Ludo; van Steensel, Bas; Myler, Peter J.; Borst, Piet (August 2012). "Glucosylated Hydroxymethyluracil, DNA Base J, Prevents Transcriptional Readthrough in Leishmania". Cell. 150 (5): 909–921. doi:10.1016/j.cell.2012.07.030. PMC 3684241. PMID 22939620.
- ^ Hazelbaker, Dane Z.; Buratowski, Stephen (November 2012). "Transcription: Base J Blocks the Way". Current Biology. 22 (22): R960–R962. doi:10.1016/j.cub.2012.10.010. PMC 3648658. PMID 23174300.
