Lanosterol synthase
| lanosterol synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 5.4.99.7 | ||||||||
| CAS no. | 9032-71-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| LSS | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | LSS, OSC, CTRCT44, lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), lanosterol synthase, HYPT14, APMR4 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 600909 MGI: 1336155 HomoloGene: 37408 GeneCards: LSS | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Lanosterol synthase (EC 5.4.99.7) is an oxidosqualene cyclase (OSC) enzyme that converts (S)-2,3-oxidosqualene to a protosterol cation and finally to lanosterol.[5] Lanosterol is a key four-ringed intermediate in cholesterol biosynthesis.[6][7] In humans, lanosterol synthase is encoded by the LSS gene.[8][9]
In eukaryotes, lanosterol synthase is an integral monotopic protein associated with the cytosolic side of the endoplasmic reticulum.[10] Some evidence suggests that the enzyme is a soluble, non-membrane bound protein in the few prokaryotes that produce it.[11]
Due to the enzyme's role in cholesterol biosynthesis, there is interest in lanosterol synthase inhibitors as potential cholesterol-reducing drugs, to complement existing statins.[12]
Mechanism[edit]
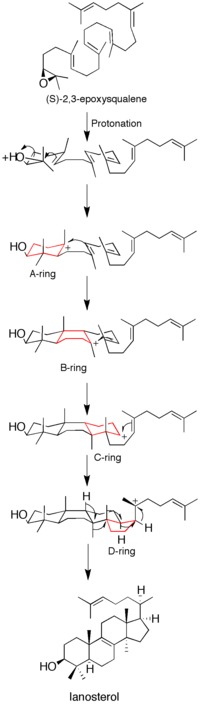
Though some data on the mechanism has been obtained by the use of suicide inhibitors, mutagenesis studies, and homology modeling, it is still not fully understood how the enzyme catalyzes the formation of lanosterol.[12]
Initial epoxide protonation and ring opening[edit]
Before the acquisition of the protein's X-ray crystal structure, site-directed mutagenesis was used to determine residues key to the enzyme's catalytic activity. It was determined that an aspartic acid residue (D455) and two histidine residues (H146 and H234) were essential to enzyme function. Corey et al. hypothesized that the aspartic acid acts by protonating the substrate's epoxide ring, thus increasing its susceptibility to intramolecular attack by the nearest double bond, with H146 possibly intensifying the proton donor ability of the aspartic acid through hydrogen bonding.[13] After acquisition of the X-ray crystal structure of the enzyme, the role of D455 as a proton donor to the substrate's epoxide was confirmed, though it was found that D455 is more likely stabilized by hydrogen bonding from two cysteine residues (C456 and C533) than from the earlier suggested histidine.[12]
Ring formation cascade[edit]
Epoxide protonation activates the substrate, setting off a cascade of ring forming reactions. Four rings in total (A through D) are formed, producing the cholesterol backbone.[12] Though the idea of a concerted formation of all four rings had been entertained in the past, kinetic studies with (S)-2,3-oxidosqualene analogs showed that product formation is achieved through discrete carbocation intermediates (see Figure 1). Isolation of monocyclic and bicyclic products from lanosterol synthase mutants has further weakened the hypothesis of a concerted mechanism.[14][15] Evidence suggests that epoxide ring opening and A ring formation is concerted, though.[16]
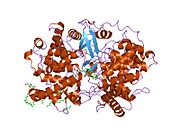
Structure[edit]
Lanosterol synthase is a two-domain monomeric protein[10] composed of two connected (α/α) barrel domains and three smaller β-structures. The enzyme active site is in the center of the protein, closed off by a constricted channel. Passage of the (S)-2,3-epoxysqualene substrate through the channel requires a change in protein conformation. In eukaryotes, a hydrophobic surface (6% of the total enzyme surface area) is the ER membrane-binding region (see Figure 2).[12]
The enzyme contains five fingerprint regions containing Gln-Trp motifs, which are also present in the highly analogous bacterial enzyme squalene-hopene cyclase.[12] Residues of these fingerprint regions contain stacked sidechains which are thought to contribute to enzyme stability during the highly exergonic cyclization reactions catalyzed by the enzyme.[17]
Function[edit]
Catalysis of lanosterol formation[edit]
Lanosterol synthase catalyzes the conversion of (S)-2,3-epoxysqualene to lanosterol, a key four-ringed intermediate in cholesterol biosynthesis.[6][7] Thus, it in turn provides the precursor to estrogens, androgens, progestogens, glucocorticoids, mineralocorticoids, and neurosteroids. In eukaryotes the enzyme is bound to the cytosolic side of the endoplasmic reticulum membrane.[10] While cholesterol synthesis is mostly associated with eukaryotes, few prokaryotes have been found to express lanosterol synthase; it has been found as a soluble protein in Methylococcus capsulatus.[11]
Catalysis of epoxylanosterol formation[edit]
Lanosterol synthase also catalyzes the cyclization of 2,3;22,23-diepoxysqualene to 24(S),25-epoxylanosterol,[18] which is later converted to 24(S),25-epoxycholesterol.[19] Since the enzyme affinity for this second substrate is greater than for the monoepoxy (S)-2,3-epoxysqualene, under partial inhibition conversion of 2,3;22,23-diepoxysqualene to 24(S),25-epoxylanosterol is favored over lanosterol synthesis.[20] This has relevance for disease prevention and treatment.
Clinical significance[edit]
Enzyme inhibitors as cholesterol-lowering drugs[edit]
Interest has grown in lanosterol synthase inhibitors as drugs to lower blood cholesterol and treat atherosclerosis. The widely popular statin drugs currently used to lower LDL (low-density lipoprotein) cholesterol function by inhibiting HMG-CoA reductase activity.[6] Because this enzyme catalyzes the formation of precursors far upstream of (S)-2,3-epoxysqualene and cholesterol, statins may negatively influence amounts of intermediates required for other biosynthetic pathways (e.g. synthesis of isoprenoids, coenzyme Q). Thus, lanosterol synthase, which is more closely tied to cholesterol biosynthesis than HMG-CoA reductase, is an attractive drug target.[21]
Lanosterol synthase inhibitors are thought to lower LDL and VLDL cholesterol by a dual control mechanism. Studies in which lanosterol synthase is partially inhibited have shown both a direct decrease in lanosterol formation and a decrease in HMG-CoA reductase activity. The oxysterol 24(S),25-epoxylanosterol, which is preferentially formed over lanosterol during partial lanosterol synthase inhibition, is believed to be responsible for this inhibition of HMG-CoA reductase activity.[22]
Evolution[edit]
It is believed that oxidosqualene cyclases (OSCs, the class to which lanosterol cyclase belongs) evolved from bacterial squalene-hopene cyclase (SHC), which is involved with the formation of hopanoids. Phylogenetic trees constructed from the amino acid sequences of OSCs in diverse organisms suggest a single common ancestor, and that the synthesis pathway evolved only once.[23] The discovery of steranes including cholestane in 2.7-billion year-old shales from Pilbara Craton, Australia, suggests that eukaryotes with OSCs and complex steroid machinery were present early in earth's history.[24]
References[edit]
- ^ a b c ENSG00000281289 GRCh38: Ensembl release 89: ENSG00000160285, ENSG00000281289 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000033105 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Dean PD, Ortiz de Montellano PR, Bloch K, Corey EJ (Jun 1967). "A soluble 2,3-oxidosqualene sterol cyclase". The Journal of Biological Chemistry. 242 (12): 3014–5. doi:10.1016/S0021-9258(18)99606-7. PMID 6027261.
- ^ a b c Huff MW, Telford DE (Jul 2005). "Lord of the rings--the mechanism for oxidosqualene:lanosterol cyclase becomes crystal clear". Trends in Pharmacological Sciences. 26 (7): 335–340. doi:10.1016/j.tips.2005.05.004. PMID 15951028.
- ^ a b Yamamoto S, Lin K, Bloch K (May 1969). "Some properties of the microsomal 2,3-oxidosqualene sterol cyclase". Proceedings of the National Academy of Sciences of the United States of America. 63 (1): 110–7. Bibcode:1969PNAS...63..110Y. doi:10.1073/pnas.63.1.110. PMC 534008. PMID 5257956.
- ^ Baker CH, Matsuda SP, Liu DR, Corey EJ (Aug 1995). "Molecular cloning of the human gene encoding lanosterol synthase from a liver cDNA library". Biochemical and Biophysical Research Communications. 213 (1): 154–160. doi:10.1006/bbrc.1995.2110. PMID 7639730.
- ^ Young M, Chen H, Lalioti MD, Antonarakis SE (May 1996). "The human lanosterol synthase gene maps to chromosome 21q22.3". Human Genetics. 97 (5): 620–624. doi:10.1007/BF02281872. PMID 8655142. S2CID 21051816.
- ^ a b c Ruf A, Müller F, D'Arcy B, Stihle M, Kusznir E, Handschin C, Morand OH, Thoma R (Mar 2004). "The monotopic membrane protein human oxidosqualene cyclase is active as monomer". Biochemical and Biophysical Research Communications. 315 (2): 247–254. doi:10.1016/j.bbrc.2004.01.052. PMID 14766201.
- ^ a b Lamb DC, Jackson CJ, Warrilow AG, Manning NJ, Kelly DE, Kelly SL (Aug 2007). "Lanosterol biosynthesis in the prokaryote Methylococcus capsulatus: insight into the evolution of sterol biosynthesis". Molecular Biology and Evolution. 24 (8): 1714–1721. doi:10.1093/molbev/msm090. PMID 17567593.
- ^ a b c d e f Thoma R, Schulz-Gasch T, D'Arcy B, Benz J, Aebi J, Dehmlow H, Hennig M, Stihle M, Ruf A (Nov 2004). "Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase". Nature. 432 (7013): 118–122. Bibcode:2004Natur.432..118T. doi:10.1038/nature02993. PMID 15525992. S2CID 364281.
- ^ Corey EJ, Cheng CH, Baker CH, Matsuda SP, Li D, Song X (February 1997). "Studies on the Substrate Binding Segments and Catalytic Action of Lanosterol Synthase. Affinity Labeling with Carbocations Derived from Mechanism-Based Analogs of 2, 3-Oxidosqualene and Site-Directed Mutagenesis Probes". J. Am. Chem. Soc. 119 (6): 1289–96. doi:10.1021/ja963228o.
- ^ Wu TK, Wang TT, Chang CH, Liu YT, Shie WS (Nov 2008). "Importance of Saccharomyces cerevisiae oxidosqualene-lanosterol cyclase tyrosine 707 residue for chair-boat bicyclic ring formation and deprotonation reactions". Organic Letters. 10 (21): 4959–62. doi:10.1021/ol802036c. PMID 18842050.
- ^ Joubert BM, Hua L, Matsuda SP (Feb 2000). "Steric bulk at position 454 in Saccharomyces cerevisiae lanosterol synthase influences B-ring formation but not deprotonation". Organic Letters. 2 (3): 339–41. doi:10.1021/ol9912940. PMID 10814317.
- ^ Corey EJ, Cheng CH, Baker CH, Matsuda SP, Li D, Song X (February 1997). "Methodology for the Preparation of Pure Recombinant S. cerevisiae Lanosterol Synthase Using a Baculovirus Expression System. Evidence That Oxirane Cleavage and A-Ring Formation Are Concerted in the Biosynthesis of Lanosterol from 2,3-Oxidosqualene". J. Am. Chem. Soc. 119 (6): 1277–88. doi:10.1021/ja963227w.
- ^ Wendt KU, Poralla K, Schulz GE (Sep 1997). "Structure and function of a squalene cyclase". Science. 277 (5333): 1811–15. doi:10.1126/science.277.5333.1811. PMID 9295270.
- ^ Corey EJ, Gross SK (Aug 1967). "Formation of sterols by the action of 2,3-oxidosqualene-sterol cyclase on the factitious substrates 2,3:22,23-dioxidosqualene and 2,3-oxido-22,23-dihydrosqualene". Journal of the American Chemical Society. 89 (17): 4561–2. doi:10.1021/ja00993a079. PMID 6046552.
- ^ Nelson JA, Steckbeck SR, Spencer TA (Feb 1981). "Biosynthesis of 24,25-epoxycholesterol from squalene 2,3;22,23-dioxide". The Journal of Biological Chemistry. 256 (3): 1067–8. doi:10.1016/S0021-9258(19)69921-7. PMID 7451488.
- ^ Boutaud O, Dolis D, Schuber F (Oct 1992). "Preferential cyclization of 2,3(S):22(S),23-dioxidosqualene by mammalian 2,3-oxidosqualene-lanosterol cyclase". Biochemical and Biophysical Research Communications. 188 (2): 898–904. doi:10.1016/0006-291X(92)91140-L. PMID 1445330.
- ^ Telford DE, Lipson SM, Barrett PH, Sutherland BG, Edwards JY, Aebi JD, Dehmlow H, Morand OH, Huff MW (Dec 2005). "A novel inhibitor of oxidosqualene:lanosterol cyclase inhibits very low-density lipoprotein apolipoprotein B100 (apoB100) production and enhances low-density lipoprotein apoB100 catabolism through marked reduction in hepatic cholesterol content". Arteriosclerosis, Thrombosis, and Vascular Biology. 25 (12): 2608–14. doi:10.1161/01.ATV.0000189158.28455.94. PMID 16210564.
- ^ Panini SR, Gupta A, Sexton RC, Parish EJ, Rudney H (Oct 1987). "Regulation of sterol biosynthesis and of 3-hydroxy-3-methylglutaryl-coenzyme A reductase activity in cultured cells by progesterone". The Journal of Biological Chemistry. 262 (30): 14435–40. doi:10.1016/S0021-9258(18)47813-1. PMID 3667583.
- ^ Pearson A, Budin M, Brocks JJ (Dec 2003). "Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus". Proceedings of the National Academy of Sciences of the United States of America. 100 (26): 15352–7. Bibcode:2003PNAS..10015352P. doi:10.1073/pnas.2536559100. PMC 307571. PMID 14660793.
- ^ Brocks JJ, Logan GA, Buick R, Summons RE (Aug 1999). "Archean molecular fossils and the early rise of eukaryotes". Science. 285 (5430): 1033–6. Bibcode:1999Sci...285.1033B. CiteSeerX 10.1.1.516.9123. doi:10.1126/science.285.5430.1033. PMID 10446042.
Further reading[edit]
- Roessler E, Mittaz L, Du Y, Scott HS, Chang J, Rossier C, Guipponi M, Matsuda SP, Muenke M, Antonarakis SE (Nov 1999). "Structure of the human Lanosterol synthase gene and its analysis as a candidate for holoprosencephaly (HPE1)". Human Genetics. 105 (5): 489–95. doi:10.1007/s004390051135 (inactive 2024-04-03). PMID 10598817.
{{cite journal}}: CS1 maint: DOI inactive as of April 2024 (link) - Ma J, Dempsey AA, Stamatiou D, Marshall KW, Liew CC (Mar 2007). "Identifying leukocyte gene expression patterns associated with plasma lipid levels in human subjects". Atherosclerosis. 191 (1): 63–72. doi:10.1016/j.atherosclerosis.2006.05.032. PMID 16806233.
- Beyea MM, Heslop CL, Sawyez CG, Edwards JY, Markle JG, Hegele RA, Huff MW (Feb 2007). "Selective up-regulation of LXR-regulated genes ABCA1, ABCG1, and APOE in macrophages through increased endogenous synthesis of 24(S),25-epoxycholesterol". The Journal of Biological Chemistry. 282 (8): 5207–16. doi:10.1074/jbc.M611063200. PMID 17186944.
- Dang H, Liu Y, Pang W, Li C, Wang N, Shyy JY, Zhu Y (Mar 2009). "Suppression of 2,3-oxidosqualene cyclase by high fat diet contributes to liver X receptor-alpha-mediated improvement of hepatic lipid profile". The Journal of Biological Chemistry. 284 (10): 6218–26. doi:10.1074/jbc.M803702200. PMC 2649084. PMID 19119143.
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T, Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai K, Isogai T, Sugano S (Jan 2006). "Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes". Genome Research. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- Sung CK, Shibuya M, Sankawa U, Ebizuka Y (Oct 1995). "Molecular cloning of cDNA encoding human lanosterol synthase". Biological & Pharmaceutical Bulletin. 18 (10): 1459–61. doi:10.1248/bpb.18.1459. PMID 8593458.
- Young M, Chen H, Lalioti MD, Antonarakis SE (May 1996). "The human lanosterol synthase gene maps to chromosome 21q22.3". Human Genetics. 97 (5): 620–4. doi:10.1007/BF02281872. PMID 8655142. S2CID 21051816.
- Peffley DM, Gayen AK, Morand OH (Aug 1998). "Down-regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase mRNA levels and synthesis in syrian hamster C100 cells by the oxidosqualene cyclase inhibitor [4'-(6-allyl-ethyl-amino-hexyloxy)-2'-fluoro-phenyl]-(4-bromophenyl)-me thanone (Ro 48-8071): comparison to simvastatin". Biochemical Pharmacology. 56 (4): 439–49. doi:10.1016/S0006-2952(98)00083-5. PMID 9763219.
- Lu Y, Dollé ME, Imholz S, van 't Slot R, Verschuren WM, Wijmenga C, Feskens EJ, Boer JM (Dec 2008). "Multiple genetic variants along candidate pathways influence plasma high-density lipoprotein cholesterol concentrations". Journal of Lipid Research. 49 (12): 2582–9. doi:10.1194/jlr.M800232-JLR200. PMID 18660489.
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR (May 2005). "Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution". Science. 308 (5725): 1149–54. Bibcode:2005Sci...308.1149C. doi:10.1126/science.1108625. PMID 15790807. S2CID 13047538.
- Pyrah IT, Kalinowski A, Jackson D, Davies W, Davis S, Aldridge A, Greaves P (2001). "Toxicologic lesions associated with two related inhibitors of oxidosqualene cyclase in the dog and mouse". Toxicologic Pathology. 29 (2): 174–9. doi:10.1080/019262301317052440. PMID 11421484. S2CID 38721319.
- Bonaldo MF, Lennon G, Soares MB (Sep 1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Research. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Ruf A, Müller F, D'Arcy B, Stihle M, Kusznir E, Handschin C, Morand OH, Thoma R (Mar 2004). "The monotopic membrane protein human oxidosqualene cyclase is active as monomer". Biochemical and Biophysical Research Communications. 315 (2): 247–54. doi:10.1016/j.bbrc.2004.01.052. PMID 14766201.
- Mark M, Muller P, Maier R, Eisele B (Jan 1996). "Effects of a novel 2,3-oxidosqualene cyclase inhibitor on the regulation of cholesterol biosynthesis in HepG2 cells". Journal of Lipid Research. 37 (1): 148–58. doi:10.1016/S0022-2275(20)37643-4. PMID 8820110.
External links[edit]
- Lanosterol+synthase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)