Neuropilin
| neuropilin 1 | |||||||
|---|---|---|---|---|---|---|---|
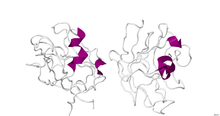 Crystallographic structure of the dimeric B1 domain of human neuropilin 1.[1] | |||||||
| Identifiers | |||||||
| Symbol | NRP1 | ||||||
| NCBI gene | 8829 | ||||||
| HGNC | 8004 | ||||||
| OMIM | 602069 | ||||||
| PDB | 3I97 | ||||||
| RefSeq | NM_001024628 | ||||||
| UniProt | O14786 | ||||||
| Other data | |||||||
| Locus | Chr. 10 p12 | ||||||
| |||||||
| neuropilin 2 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | NRP2 | ||||||
| NCBI gene | 8828 | ||||||
| HGNC | 8005 | ||||||
| OMIM | 602070 | ||||||
| RefSeq | NM_201279 | ||||||
| UniProt | O60462 | ||||||
| Other data | |||||||
| Locus | Chr. 2 q34 | ||||||
| |||||||
Neuropilin is a protein receptor active in neurons.
There are two forms of Neuropilins, NRP-1 and NRP-2. They are transmembrane glycoproteins, and predominantly co-receptors for another class of proteins known as semaphorins. Of the semaphorins, NRP-1 and NRP-2 are specifically receptors for class-3 semaphorins, which, among many things, are responsible for axon guidance during the development of the nervous system in vertebrates.
Neuropilins work as co-receptors as they have a very small cytoplasmic domain and thus rely upon other molecules (normally plexins) to transduce their signals across a cell membrane. Neuropilins generally work as dimers and different combinations have different affinities for molecules. For example, NRP-1 homodimers have high affinity for Sema3A, whilst NRP-2 homodimers have high affinity for Sema3F. Another ligand for neuropilins is VEGF, a growth factor involved in the regulation of angiogenesis.
The pleiotropic nature of the NRP receptors results in their involvement in multiple signalling pathways, such as axon guidance and angiogenesis, the immune response and remyelination.[2]
Applications
Neuropilin-1 is a therapeutic target protein in the treatment for leukemia and lymphoma, since It has been shown that there is increased expression in neuropilin-1 in leukemia and lymphoma cell lines.[3] Also, antagonism of neuropilin-1 has been found to inhibit tumour cell migration and adhesion.[4]
Structure
Neuropilins contain the following four domains:
- N-terminal CUB domain (for complement C1r/C1s, Uegf, Bmp1)
- Coagulation factor 5/8 type, C-terminal (discoidin domain)
- MAM domain (for meprin, A-5 protein, and receptor protein-tyrosine phosphatase mu)
- C-terminal neuropilin
The structure of B1 domain (coagulation factor 5/8 type) of neuropilin-1 was determined through X-Ray Diffraction with a resolution of 2.90 Å. The secondary structure of this domain is 5% alpha helical and 46% beta sheet.[1]
Ramachandran plot.[5]
References
- ^ a b PDB: 3I97; Jarvis A, Allerston CK, Jia H, Herzog B, Garza-Garcia A, Winfield N, Ellard K, Aqil R, Lynch R, Chapman C, Hartzoulakis B, Nally J, Stewart M, Cheng L, Menon M, Tickner M, Djordjevic S, Driscoll PC, Zachary I, Selwood DL (March 2010). "Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction". J. Med. Chem. 53 (5): 2215–26. doi:10.1021/jm901755g. PMC 2841442. PMID 20151671.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mecollari, V; Nieuwenhuis, B; Verhaagen, J (2014). "A perspective on the role of class III semaphorin signaling in central nervous system trauma". Frontiers in Cellular Neuroscience. 8: 328. doi:10.3389/fncel.2014.00328. PMC 4209881. PMID 25386118.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Karjalainen K, Jaalouk DE, Bueso-Ramos CE, Zurita AJ, Kuniyasu A, Eckhardt BL, Marini FC, Lichtiger B, O'Brien S, Kantarjian HM, Cortes JE, Koivunen E, Arap W, Pasqualini R (November 2010). "Targeting neuropilin-1 in human leukemia and lymphoma". Blood. 117 (3): 920–927. doi:10.1182/blood-2010-05-282921. PMC 3298438. PMID 21063027.
{{cite journal}}: Cite has empty unknown parameter:|issn.=(help)CS1 maint: multiple names: authors list (link) - ^ Jia H, Cheng L, Tickner M, Bagherzadeh A, Selwood D, Zachary I (February 2010). "Neuropilin-1 antagonism in human carcinoma cells inhibits migration and enhances chemosensitivity". Br. J. Cancer. 102 (3): 541–52. doi:10.1038/sj.bjc.6605539. PMC 2822953. PMID 20087344.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "MolProbity Ramachandran analysis of PDB structure 3I97" (PDF). www.pdb.org.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
External links
- Neuropilins at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
