Siah interacting protein N-terminal domain
| Siah-Interacting Protein, N terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
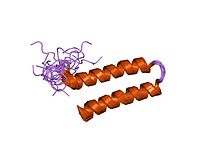 NMR structure of N-terminal domain (residues 1-77) of Siah-interacting protein. | |||||||||
| Identifiers | |||||||||
| Symbol | Siah-Interact_N | ||||||||
| Pfam | PF09032 | ||||||||
| InterPro | IPR015120 | ||||||||
| |||||||||
In molecular biology the protein domain, Siah interacting protein N-terminal domain is found at the N-terminal of the protein, Siah interacting protein (SIP). It has a helical hairpin structure with a hydrophobic core which is further stabilised by an arrangement of side chains contributed by the two amphipathic helices. The function of this domain remains to be fully elucidated, but it is known to be vital for interactions with Siah. It has also been hypothesised that SIP can dimerise through this N-terminal domain.[1]
Function
[edit]SIP protein
[edit]The SIP protein has a role as an adaptor protein, it links the E3 ubiquitin ligase activity of Siah-1 with Skp1 and Ebi F-Box protein in the degradation of beta-catenin, a transcriptional activator of TCF/LEF genes. This is important for signalling that the protein needs to undergo proteolysis at the 26S proteasome.[2]
N-terminal domain of SIP protein
[edit]More specifically, the N-terminal domain of the SIP protein is a dimerisation domain.[2] Its precise function is yet to be elucidated. More recent studies have shown that when the N-terminal domain is shortened, or in other words truncated by a nonsense mutation, it results in an increase in the import of SIP into the nucleus and enhances its proapoptotic effect (programmed cell death).[3] These findings indicate that the SIP protein and in particular the N-terminal domain may provide important information about drug resistance.
Structure
[edit]The N terminal domain is a dimer. Each monomer has an alpha helical hairpin in which the two alpha helices are connected by a tight 3-residue turn. Hairpins from two monomers associate as a four-helix bundle.[2]
References
[edit]- ^ Bhattacharya S, Lee YT, Michowski W, Jastrzebska B, Filipek A, Kuznicki J, Chazin WJ (July 2005). "The modular structure of SIP facilitates its role in stabilizing multiprotein assemblies". Biochemistry. 44 (27): 9462–71. doi:10.1021/bi0502689. PMID 15996101.
- ^ a b c Santelli E, Leone M, Li C, Fukushima T, Preece NE, Olson AJ, et al. (2005). "Structural analysis of Siah1-Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex". J Biol Chem. 280 (40): 34278–87. doi:10.1074/jbc.M506707200. PMID 16085652.
- ^ Luo J, Yang J, Yu BY, Liu W, Li M, Zhuang SM (2010). "Identification of Siah-interacting protein as a potential regulator of apoptosis and curcumin resistance". Oncogene. 29 (48): 6357–66. doi:10.1038/onc.2010.358. PMID 20729913.
