Reference genome: Difference between revisions
Aarón Mayo (talk | contribs) No edit summary |
Aarón Mayo (talk | contribs) Move information about sequencing cost and add new subsection about reference genome limitations |
||
| Line 1: | Line 1: | ||
[[File:Wellcome genome bookcase.png|thumb|right|250px|The first printout of the human reference genome presented as a series of books, displayed at the [[Wellcome Collection]], London]] |
[[File:Wellcome genome bookcase.png|thumb|right|250px|The first printout of the human reference genome presented as a series of books, displayed at the [[Wellcome Collection]], London]] |
||
A '''reference genome''' (also known as a '''reference assembly''') is a digital [[nucleic acid sequence]] database, assembled by scientists as a representative example of the [[genome|set of genes]] in one idealized individual organism of a species. As they are assembled from the sequencing of [[DNA]] from a number of individual donors, reference [[genome]]s do not accurately represent the set of genes of any single individual organism. Instead a reference provides a [[haploid]] mosaic of different DNA sequences from each donor. For example, the most recent human reference genome (assembly ''[[GRCh38|GRCh38/hg38]]'') is derived from >60 [[genomic library|genomic clone libraries]]. <ref name=GRC_FAQ>{{cite web |title=How many individuals were sequenced for the human reference genome assembly? |url=https://www.ncbi.nlm.nih.gov/grc/help/faq/#human-reference-genome-individuals |website=Genome Reference Consortium |access-date=7 April 2022}}</ref> There are reference genomes for multiple species of [[viruses]], [[bacteria]], [[fungus]], [[plants]], and [[animals]]. |
A '''reference genome''' (also known as a '''reference assembly''') is a digital [[nucleic acid sequence]] database, assembled by scientists as a representative example of the [[genome|set of genes]] in one idealized individual organism of a species. As they are assembled from the sequencing of [[DNA]] from a number of individual donors, reference [[genome]]s do not accurately represent the set of genes of any single individual organism. Instead a reference provides a [[haploid]] mosaic of different DNA sequences from each donor. For example, the most recent human reference genome (assembly ''[[GRCh38|GRCh38/hg38]]'') is derived from >60 [[genomic library|genomic clone libraries]]. <ref name=GRC_FAQ>{{cite web |title=How many individuals were sequenced for the human reference genome assembly? |url=https://www.ncbi.nlm.nih.gov/grc/help/faq/#human-reference-genome-individuals |website=Genome Reference Consortium |access-date=7 April 2022}}</ref> There are reference genomes for multiple species of [[viruses]], [[bacteria]], [[fungus]], [[plants]], and [[animals]]. Reference genomes are typically used as a guide on which new genomes are built, enabling them to be assembled much more quickly and cheaply than the initial [[Human Genome Project]]. Reference genomes can be accessed online at several locations, using dedicated browsers such as [[Ensembl]] or [[UCSC Genome Browser]].<ref name="ensembl">{{cite journal | vauthors = Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Gräf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kähäri A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Slater G, Smedley D, Spudich G, Trevanion S, Vilella AJ, Vogel J, White S, Wood M, Birney E, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Kasprzyk A, Proctor G, Smith J, Ureta-Vidal A, Searle S | display-authors = 6 | title = Ensembl 2008 | journal = Nucleic Acids Research | volume = 36 | issue = Database issue | pages = D707-D714 | date = January 2008 | pmid = 18000006 | pmc = 2238821 | doi = 10.1093/nar/gkm988 }}</ref> |
||
| ⚫ | As the cost of [[DNA sequencing]] falls, and new [[full genome sequencing]] technologies emerge, more genome sequences continue to be generated. |
||
Reference genomes can be accessed online at several locations, using dedicated browsers such as [[Ensembl]] or [[UCSC Genome Browser]].<ref name="ensembl">{{cite journal | vauthors = Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Gräf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kähäri A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Slater G, Smedley D, Spudich G, Trevanion S, Vilella AJ, Vogel J, White S, Wood M, Birney E, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Kasprzyk A, Proctor G, Smith J, Ureta-Vidal A, Searle S | display-authors = 6 | title = Ensembl 2008 | journal = Nucleic Acids Research | volume = 36 | issue = Database issue | pages = D707-D714 | date = January 2008 | pmid = 18000006 | pmc = 2238821 | doi = 10.1093/nar/gkm988 }}</ref> |
|||
==Properties of reference genomes== |
==Properties of reference genomes== |
||
| Line 21: | Line 17: | ||
===Human reference genome=== |
===Human reference genome=== |
||
The original human reference genome was derived from thirteen anonymous volunteers from [[Buffalo, New York]]. Donors were recruited by advertisement in ''[[The Buffalo News]]'', on Sunday, March 23, 1997. The first ten male and ten female volunteers were invited to make an appointment with the project's [[genetic counselors]] and donate blood from which DNA was extracted. As a result of how the DNA samples were processed, about 80 percent of the reference genome came from eight people and one male, designated ''RP11'', accounts for 66 percent of the total. The [[ABO blood group system]] differs among humans, but the human reference genome contains only an [[ABO (gene)|O allele]], although the others are [[Genome annotation#Genome annotation|annotated]].<ref name="Guide">{{cite book |title=A short guide to the human genome | vauthors = Scherer S |year=2008 |publisher=CSHL Press |isbn=978-0-87969-791-4 |page=135 }}</ref><ref name=Editorial>{{cite journal | vauthors = | title = E pluribus unum | journal = Nature Methods | volume = 7 | issue = 5 | pages = 331 | date = May 2010 | pmid = 20440876 | doi = 10.1038/nmeth0510-331 | doi-access = free }}</ref><ref name="Change">{{cite journal | vauthors = Ballouz S, Dobin A, Gillis JA | title = Is it time to change the reference genome? | journal = Genome Biology | volume = 20 | issue = 1 | pages = 159 | date = August 2019 | pmid = 31399121 | pmc = 6688217 | doi = 10.1186/s13059-019-1774-4 | doi-access = free }}</ref><ref name="PLOS_Rosen">{{cite journal | vauthors = Rosenfeld JA, Mason CE, Smith TM | title = Limitations of the human reference genome for personalized genomics | journal = PloS One | volume = 7 | issue = 7 | pages = e40294 | date = 11 July 2012 | pmid = 22811759 | pmc = 3394790 | doi = 10.1371/journal.pone.0040294 | doi-access = free | bibcode = 2012PLoSO...740294R }}</ref><ref name="NYT"/> |
The original human reference genome was derived from thirteen anonymous volunteers from [[Buffalo, New York]]. Donors were recruited by advertisement in ''[[The Buffalo News]]'', on Sunday, March 23, 1997. The first ten male and ten female volunteers were invited to make an appointment with the project's [[genetic counselors]] and donate blood from which DNA was extracted. As a result of how the DNA samples were processed, about 80 percent of the reference genome came from eight people and one male, designated ''RP11'', accounts for 66 percent of the total. The [[ABO blood group system]] differs among humans, but the human reference genome contains only an [[ABO (gene)|O allele]], although the others are [[Genome annotation#Genome annotation|annotated]].<ref name="Guide">{{cite book |title=A short guide to the human genome | vauthors = Scherer S |year=2008 |publisher=CSHL Press |isbn=978-0-87969-791-4 |page=135 }}</ref><ref name=Editorial>{{cite journal | vauthors = | title = E pluribus unum | journal = Nature Methods | volume = 7 | issue = 5 | pages = 331 | date = May 2010 | pmid = 20440876 | doi = 10.1038/nmeth0510-331 | doi-access = free }}</ref><ref name="Change">{{cite journal | vauthors = Ballouz S, Dobin A, Gillis JA | title = Is it time to change the reference genome? | journal = Genome Biology | volume = 20 | issue = 1 | pages = 159 | date = August 2019 | pmid = 31399121 | pmc = 6688217 | doi = 10.1186/s13059-019-1774-4 | doi-access = free }}</ref><ref name="PLOS_Rosen">{{cite journal | vauthors = Rosenfeld JA, Mason CE, Smith TM | title = Limitations of the human reference genome for personalized genomics | journal = PloS One | volume = 7 | issue = 7 | pages = e40294 | date = 11 July 2012 | pmid = 22811759 | pmc = 3394790 | doi = 10.1371/journal.pone.0040294 | doi-access = free | bibcode = 2012PLoSO...740294R }}</ref><ref name="NYT"/> |
||
[[File:Cost per Genome.png|thumb|444x444px|Evolution of the cost of sequencing a human genome from 2001 to 2021]] |
|||
| ⚫ | As the cost of [[DNA sequencing]] falls, and new [[full genome sequencing]] technologies emerge, more genome sequences continue to be generated. In several cases people such as [[James D. Watson]] had their genome assembled using [[Massive parallel sequencing|massive parallel DNA sequencing]].<ref name="Watson" /><ref>The exception to this is [[J. Craig Venter]] whose DNA was sequenced and assembled using [[shotgun sequencing]] methods.</ref> Comparison between the reference (assembly NCBI36/hg18) and Watson's genome revealed 3.3 million [[single nucleotide polymorphism]] differences, while about 1.4 percent of his DNA could not be matched to the reference genome at all.<ref name="NYT">{{cite news | title=Genome of DNA Pioneer Is Deciphered | vauthors = Wade N | work=New York Times | date=May 31, 2007 | access-date=February 21, 2009 | url=https://www.nytimes.com/2007/05/31/science/31cnd-gene.html}}</ref><ref name="Watson">{{cite journal | vauthors = Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM | display-authors = 6 | title = The complete genome of an individual by massively parallel DNA sequencing | journal = Nature | volume = 452 | issue = 7189 | pages = 872–876 | date = April 2008 | pmid = 18421352 | doi = 10.1038/nature06884 | doi-access = free | bibcode = 2008Natur.452..872W }}</ref> For regions where there is known to be large-scale variation, sets of alternate [[Locus (genetics)|loci]] are assembled alongside the reference locus. |
||
[[File:Human genome assembly GRCh38 chromosomes ideogram NCBI.png|thumb|496x496px|Chromosomes ideogram of the human reference genome assembly GRCh38/hg38. Characteristic bands patterns are displayed in black, grey and white, while the gaps and partially assembled regions are displayed in blue and rose, respectively.Reference: Genome Data Viewer of the NCBI.<ref>{{Cite web |title=Genome Data Viewer - NCBI |url=https://www.ncbi.nlm.nih.gov/genome/gdv/browser/genome/?id=GCF_000001405.40 |access-date=2022-08-18 |website=www.ncbi.nlm.nih.gov}}</ref>]] |
[[File:Human genome assembly GRCh38 chromosomes ideogram NCBI.png|thumb|496x496px|Chromosomes ideogram of the human reference genome assembly GRCh38/hg38. Characteristic bands patterns are displayed in black, grey and white, while the gaps and partially assembled regions are displayed in blue and rose, respectively.Reference: Genome Data Viewer of the NCBI.<ref>{{Cite web |title=Genome Data Viewer - NCBI |url=https://www.ncbi.nlm.nih.gov/genome/gdv/browser/genome/?id=GCF_000001405.40 |access-date=2022-08-18 |website=www.ncbi.nlm.nih.gov}}</ref>]] |
||
The latest patch of the human reference genome released by the [[Genome Reference Consortium]] was GRCh38 in 2017.<ref>{{cite journal | vauthors = Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, Fulton RS, Kremitzki M, Magrini V, Markovic C, McGrath S, Steinberg KM, Auger K, Chow W, Collins J, Harden G, Hubbard T, Pelan S, Simpson JT, Threadgold G, Torrance J, Wood JM, Clarke L, Koren S, Boitano M, Peluso P, Li H, Chin CS, Phillippy AM, Durbin R, Wilson RK, Flicek P, Eichler EE, Church DM | display-authors = 6 | title = Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly | journal = Genome Research | volume = 27 | issue = 5 | pages = 849–864 | date = May 2017 | pmid = 28396521 | pmc = 5411779 | doi = 10.1101/gr.213611.116 }}</ref> This build only has 120 gaps across all the assembly, which implies a great improvement in comparison with previous genome assemblies; the first version had roughly 150,000 gaps.<ref name="Editorial" /> It presents gaps mostly in areas concerning [[Telomere|telomeres]], [[Centromere|centromeres]] and long [[Repeated sequence (DNA)|repetitive sequences]], being the biggest gap along the long arm of the Y chromosome, a region of ~30 Mb length (~52% of the Y chromosome length).<ref>{{Cite web |title=GRCh38.p14 - hg38 - Genome - Assembly - NCBI - Statistics Report |url=https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.40 |access-date=2022-08-18 |website=www.ncbi.nlm.nih.gov}}</ref> The number of [[genomic library|genomic clone libraries]] contributing to the reference has increased steadily to >60 along the years, although individual ''RP11'' still accounts for 70% of the genome. <ref name="GRC_FAQ" /> Genomic analysis of this anonymous male suggests that he is of African-European ancestry. <ref name="GRC_FAQ" /> In 2022, the Telomere-to-Telomere (T2T) Consortium<ref>{{Cite web |title=Telomere-to-Telomere |url=https://www.genome.gov/about-genomics/telomere-to-telomere |access-date=2022-08-16 |website=NHGRI |language=en}}</ref> published the first completely assembled reference genome (version T2T-CHM13), without any gaps in the assembly.<ref>{{cite journal | vauthors = Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng H, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PG, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AF, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JM, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O'Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM | display-authors = 6 | title = The complete sequence of a human genome | journal = Science | volume = 376 | issue = 6588 | pages = 44–53 | date = April 2022 | pmid = 35357919 | pmc = 9186530 | doi = 10.1126/science.abj6987 | s2cid = 247854936 | bibcode = 2022Sci...376...44N }}</ref><ref>{{Cite web |title=T2T-CHM13v2.0 - Genome - Assembly - NCBI |url=https://www.ncbi.nlm.nih.gov/assembly/GCF_009914755.1/ |access-date=2022-08-16 |website=www.ncbi.nlm.nih.gov}}</ref> On the other hand, according to the GRC website, their next assembly release for the human genome (version GRCh39) is currently "indefinitely postponed".<ref name=":1">{{Cite web |title=Genome Reference Consortium |url=https://www.ncbi.nlm.nih.gov/grc |access-date=2022-08-18 |website=www.ncbi.nlm.nih.gov}}</ref> |
The latest patch of the human reference genome released by the [[Genome Reference Consortium]] was GRCh38 in 2017.<ref>{{cite journal | vauthors = Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, Fulton RS, Kremitzki M, Magrini V, Markovic C, McGrath S, Steinberg KM, Auger K, Chow W, Collins J, Harden G, Hubbard T, Pelan S, Simpson JT, Threadgold G, Torrance J, Wood JM, Clarke L, Koren S, Boitano M, Peluso P, Li H, Chin CS, Phillippy AM, Durbin R, Wilson RK, Flicek P, Eichler EE, Church DM | display-authors = 6 | title = Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly | journal = Genome Research | volume = 27 | issue = 5 | pages = 849–864 | date = May 2017 | pmid = 28396521 | pmc = 5411779 | doi = 10.1101/gr.213611.116 }}</ref> This build only has 120 gaps across all the assembly, which implies a great improvement in comparison with previous genome assemblies; the first version had roughly 150,000 gaps.<ref name="Editorial" /> It presents gaps mostly in areas concerning [[Telomere|telomeres]], [[Centromere|centromeres]] and long [[Repeated sequence (DNA)|repetitive sequences]], being the biggest gap along the long arm of the Y chromosome, a region of ~30 Mb length (~52% of the Y chromosome length).<ref>{{Cite web |title=GRCh38.p14 - hg38 - Genome - Assembly - NCBI - Statistics Report |url=https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.40 |access-date=2022-08-18 |website=www.ncbi.nlm.nih.gov}}</ref> The number of [[genomic library|genomic clone libraries]] contributing to the reference has increased steadily to >60 along the years, although individual ''RP11'' still accounts for 70% of the genome. <ref name="GRC_FAQ" /> Genomic analysis of this anonymous male suggests that he is of African-European ancestry. <ref name="GRC_FAQ" /> In 2022, the Telomere-to-Telomere (T2T) Consortium<ref>{{Cite web |title=Telomere-to-Telomere |url=https://www.genome.gov/about-genomics/telomere-to-telomere |access-date=2022-08-16 |website=NHGRI |language=en}}</ref> published the first completely assembled reference genome (version T2T-CHM13), without any gaps in the assembly.<ref>{{cite journal | vauthors = Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng H, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PG, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AF, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JM, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O'Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM | display-authors = 6 | title = The complete sequence of a human genome | journal = Science | volume = 376 | issue = 6588 | pages = 44–53 | date = April 2022 | pmid = 35357919 | pmc = 9186530 | doi = 10.1126/science.abj6987 | s2cid = 247854936 | bibcode = 2022Sci...376...44N }}</ref><ref>{{Cite web |title=T2T-CHM13v2.0 - Genome - Assembly - NCBI |url=https://www.ncbi.nlm.nih.gov/assembly/GCF_009914755.1/ |access-date=2022-08-16 |website=www.ncbi.nlm.nih.gov}}</ref> On the other hand, according to the GRC website, their next assembly release for the human genome (version GRCh39) is currently "indefinitely postponed".<ref name=":1">{{Cite web |title=Genome Reference Consortium |url=https://www.ncbi.nlm.nih.gov/grc |access-date=2022-08-18 |website=www.ncbi.nlm.nih.gov}}</ref> |
||
| ⚫ | |||
Recent genome assemblies are as follows:<ref name=":0">{{cite web|url=https://genome.ucsc.edu/FAQ/FAQreleases.html#release1|title=UCSC Genome Bioinformatics: FAQ|website=genome.ucsc.edu|access-date=2016-08-18}}</ref> |
Recent genome assemblies are as follows:<ref name=":0">{{cite web|url=https://genome.ucsc.edu/FAQ/FAQreleases.html#release1|title=UCSC Genome Bioinformatics: FAQ|website=genome.ucsc.edu|access-date=2016-08-18}}</ref> |
||
| Line 61: | Line 57: | ||
|hg16 |
|hg16 |
||
|} |
|} |
||
==== Limitations ==== |
|||
For much of a genome, the reference provides a good approximation of the DNA of any single individual. But in regions with high [[gene pool|allelic diversity]], such as the [[major histocompatibility complex]] in humans and the [[major urinary proteins]] of mice, the reference genome may differ significantly from other individuals.<ref name="MHCsc">{{cite journal | author = MHC Sequencing Consortium | title = Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium | journal = Nature | volume = 401 | issue = 6756 | pages = 921–923 | date = October 1999 | pmid = 10553908 | doi = 10.1038/44853 | s2cid = 186243515 | bibcode = 1999Natur.401..921T }}</ref><ref name="Logan">{{cite journal | vauthors = Logan DW, Marton TF, Stowers L | title = Species specificity in major urinary proteins by parallel evolution | journal = PloS One | volume = 3 | issue = 9 | pages = e3280 | date = September 2008 | pmid = 18815613 | pmc = 2533699 | doi = 10.1371/journal.pone.0003280 | veditors = Vosshall LB | doi-access = free | bibcode = 2008PLoSO...3.3280L }}</ref><ref name="Hurstchapter">{{cite book |vauthors=Hurst J, Beynon RJ, Roberts SC, Wyatt TD |title=Urinary Lipocalins in Rodenta:is there a Generic Model? |series = Chemical Signals in Vertebrates 11 |publisher= Springer New York |date=October 2007 |isbn= 978-0-387-73944-1}}</ref> Due to the fact that the reference genome is a "single" distinct sequence, which gives its utility as an index or locator of genomic features, there are limitations in terms of how faithfully it represents the human genome and its [[Human genetic variation|variability]]. On the other hand, most of the samples used for reference genomes sequencing come from people of european ancestry, being these human populations the best characterized and studied at the expense of non-european populations. In 2010, it was found that, by ''de novo'' assembling genomes from african and asian populations with the NCBI reference genome (version NCBI36), these genomes had ~5Mb sequences that did not align against any region of the reference genome.<ref>{{Cite journal |last=Li |first=Ruiqiang |last2=Li |first2=Yingrui |last3=Zheng |first3=Hancheng |last4=Luo |first4=Ruibang |last5=Zhu |first5=Hongmei |last6=Li |first6=Qibin |last7=Qian |first7=Wubin |last8=Ren |first8=Yuanyuan |last9=Tian |first9=Geng |last10=Li |first10=Jinxiang |last11=Zhou |first11=Guangyu |last12=Zhu |first12=Xuan |last13=Wu |first13=Honglong |last14=Qin |first14=Junjie |last15=Jin |first15=Xin |date=January 2010 |title=Building the sequence map of the human pan-genome |url=https://www.nature.com/articles/nbt.1596 |journal=Nature Biotechnology |language=en |volume=28 |issue=1 |pages=57–63 |doi=10.1038/nbt.1596 |issn=1546-1696}}</ref> |
|||
| ⚫ | The [[1000 Genomes Project]] is creating a database to provide information about the variations in genomes across human populations, which the reference genome is not able to represent.<ref>{{cite web|url=https://www.internationalgenome.org/home |title=1000 Genomes | A Deep Catalog of Human Genetic Variation |publisher=Internationalgenome.org |date= |access-date=2022-07-19}}</ref> |
||
=== Mouse reference genome === |
=== Mouse reference genome === |
||
| Line 96: | Line 97: | ||
== Other genomes == |
== Other genomes == |
||
Since the Human Genome Project was finished, multiple international projects have started, focused on assembling reference genomes for many organisms. Model organisms (e.g., zebrafish (''[[Zebrafish|Danio rerio]]''), chicken (''[[Red junglefowl|Gallus gallus]]''), ''[[Escherichia coli]]'' etc.) are of special interest to the scientific community, as well as, for example, endangered species (e.g., Asian arowana (''[[Asian arowana|Scleropages formosus]])'' or the American bison (''[[American bison|Bison bison]]'')). As of August 2022, the NCBI database supports 71 886 partially or completely sequenced and assembled genomes from different species, such as 676 [[Mammal|mammals]], 590 [[Bird|birds]] and 865 [[Fish|fishes]]. Also noteworthy are the numbers of 1796 [[Insect|insects]] genomes, 3747 [[Fungus|fungi]], 1025 [[Plant|plants]], 33 724 [[bacteria]], 26 004 [[virus]] and 2040 [[archaea]].<ref>{{Cite web |title=Genome List - Genome - NCBI |url=https://www.ncbi.nlm.nih.gov/genome/browse#!/overview/ |access-date=2022-08-18 |website=www.ncbi.nlm.nih.gov}}</ref> A lot of these species have annotation data associated with their reference genomes that can be publicly accessed and |
Since the Human Genome Project was finished, multiple international projects have started, focused on assembling reference genomes for many organisms. Model organisms (e.g., zebrafish (''[[Zebrafish|Danio rerio]]''), chicken (''[[Red junglefowl|Gallus gallus]]''), ''[[Escherichia coli]]'' etc.) are of special interest to the scientific community, as well as, for example, endangered species (e.g., Asian arowana (''[[Asian arowana|Scleropages formosus]])'' or the American bison (''[[American bison|Bison bison]]'')). As of August 2022, the NCBI database supports 71 886 partially or completely sequenced and assembled genomes from different species, such as 676 [[Mammal|mammals]], 590 [[Bird|birds]] and 865 [[Fish|fishes]]. Also noteworthy are the numbers of 1796 [[Insect|insects]] genomes, 3747 [[Fungus|fungi]], 1025 [[Plant|plants]], 33 724 [[bacteria]], 26 004 [[virus]] and 2040 [[archaea]].<ref>{{Cite web |title=Genome List - Genome - NCBI |url=https://www.ncbi.nlm.nih.gov/genome/browse#!/overview/ |access-date=2022-08-18 |website=www.ncbi.nlm.nih.gov}}</ref> A lot of these species have annotation data associated with their reference genomes that can be publicly accessed and ''visuali''zed in genome browsers such as [[Ensembl genome database project|Ensembl]] and [[UCSC Genome Browser]].<ref>{{Cite web |title=Species List |url=https://uswest.ensembl.org/info/about/species.html |access-date=2022-08-18 |website=uswest.ensembl.org}}</ref><ref>{{Cite web |title=GenArk: UCSC Genome Archive |url=https://hgdownload.soe.ucsc.edu/hubs/ |access-date=2022-08-18 |website=hgdownload.soe.ucsc.edu}}</ref> |
||
Some examples of these international projects are: the [[Chimpanzee genome project|Chimpanzee Genome Project]], carried out between 2005 and 2013 jointly by the [[Broad Institute]] and the [[McDonnell Genome Institute]] of [[Washington University in St. Louis]], which generated the first reference genomes for 4 subspecies of ''[[Chimpanzee|Pan troglodytes]]'';<ref>{{Cite news |date=2016-03-04 |title=Chimpanzee Genome Project |language=en |work=BCM-HGSC |url=https://www.hgsc.bcm.edu/non-human-primates/chimpanzee-genome-project |access-date=2022-08-18}}</ref><ref>{{cite journal | vauthors = Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O'Connor TD, Santpere G, Cagan A, Theunert C, Casals F, Laayouni H, Munch K, Hobolth A, Halager AE, Malig M, Hernandez-Rodriguez J, Hernando-Herraez I, Prüfer K, Pybus M, Johnstone L, Lachmann M, Alkan C, Twigg D, Petit N, Baker C, Hormozdiari F, Fernandez-Callejo M, Dabad M, Wilson ML, Stevison L, Camprubí C, Carvalho T, Ruiz-Herrera A, Vives L, Mele M, Abello T, Kondova I, Bontrop RE, Pusey A, Lankester F, Kiyang JA, Bergl RA, Lonsdorf E, Myers S, Ventura M, Gagneux P, Comas D, Siegismund H, Blanc J, Agueda-Calpena L, Gut M, Fulton L, Tishkoff SA, Mullikin JC, Wilson RK, Gut IG, Gonder MK, Ryder OA, Hahn BH, Navarro A, Akey JM, Bertranpetit J, Reich D, Mailund T, Schierup MH, Hvilsom C, Andrés AM, Wall JD, Bustamante CD, Hammer MF, Eichler EE, Marques-Bonet T | display-authors = 6 | title = Great ape genetic diversity and population history | journal = Nature | volume = 499 | issue = 7459 | pages = 471–475 | date = July 2013 | pmid = 23823723 | pmc = 3822165 | doi = 10.1038/nature12228 }}</ref> the [[100K Pathogen Genome Project]], which started in 2012 with the main goal of creating a database of reference genomes for 100 000 [[pathogen]] microorganisms to use in public health, outbreaks detection, agriculture and environment;<ref>{{Cite web |title=100K Pathogen Genome Project – Genomes for Public Health & Food Safety |url=https://100kgenomes.org/ |access-date=2022-08-18 |language=en-US}}</ref> the [[Earth BioGenome Project]], which started in 2018 and aims to sequence and catalog the genomes of all the eukaryotic organisms on Earth to promote biodiversity conservation projects. Inside this big-science project there are up to 50 smaller-scale affiliated projects such as the [[Africa BioGenome Project]] or the [[1000 Fungal Genomes Project]].<ref>{{cite journal | vauthors = Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, Durbin R, Edwards SV, Forest F, Gilbert MT, Goldstein MM, Grigoriev IV, Hackett KJ, Haussler D, Jarvis ED, Johnson WE, Patrinos A, Richards S, Castilla-Rubio JC, van Sluys MA, Soltis PS, Xu X, Yang H, Zhang G | display-authors = 6 | title = Earth BioGenome Project: Sequencing life for the future of life | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 115 | issue = 17 | pages = 4325–4333 | date = April 2018 | pmid = 29686065 | pmc = 5924910 | doi = 10.1073/pnas.1720115115 }}</ref><ref>{{Cite web |title=African BioGenome Project – Genomics in the service of conservation and improvement of African biological diversity |url=https://africanbiogenome.org/ |access-date=2022-08-18 |language=en-US}}</ref><ref>{{Cite web |title=1000 Fungal Genomes Project |url=https://mycocosm.jgi.doe.gov/mycocosm/home/1000-fungal-genomes |access-date=2022-08-18 |website=mycocosm.jgi.doe.gov}}</ref> |
Some examples of these international projects are: the [[Chimpanzee genome project|Chimpanzee Genome Project]], carried out between 2005 and 2013 jointly by the [[Broad Institute]] and the [[McDonnell Genome Institute]] of [[Washington University in St. Louis]], which generated the first reference genomes for 4 subspecies of ''[[Chimpanzee|Pan troglodytes]]'';<ref>{{Cite news |date=2016-03-04 |title=Chimpanzee Genome Project |language=en |work=BCM-HGSC |url=https://www.hgsc.bcm.edu/non-human-primates/chimpanzee-genome-project |access-date=2022-08-18}}</ref><ref>{{cite journal | vauthors = Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O'Connor TD, Santpere G, Cagan A, Theunert C, Casals F, Laayouni H, Munch K, Hobolth A, Halager AE, Malig M, Hernandez-Rodriguez J, Hernando-Herraez I, Prüfer K, Pybus M, Johnstone L, Lachmann M, Alkan C, Twigg D, Petit N, Baker C, Hormozdiari F, Fernandez-Callejo M, Dabad M, Wilson ML, Stevison L, Camprubí C, Carvalho T, Ruiz-Herrera A, Vives L, Mele M, Abello T, Kondova I, Bontrop RE, Pusey A, Lankester F, Kiyang JA, Bergl RA, Lonsdorf E, Myers S, Ventura M, Gagneux P, Comas D, Siegismund H, Blanc J, Agueda-Calpena L, Gut M, Fulton L, Tishkoff SA, Mullikin JC, Wilson RK, Gut IG, Gonder MK, Ryder OA, Hahn BH, Navarro A, Akey JM, Bertranpetit J, Reich D, Mailund T, Schierup MH, Hvilsom C, Andrés AM, Wall JD, Bustamante CD, Hammer MF, Eichler EE, Marques-Bonet T | display-authors = 6 | title = Great ape genetic diversity and population history | journal = Nature | volume = 499 | issue = 7459 | pages = 471–475 | date = July 2013 | pmid = 23823723 | pmc = 3822165 | doi = 10.1038/nature12228 }}</ref> the [[100K Pathogen Genome Project]], which started in 2012 with the main goal of creating a database of reference genomes for 100 000 [[pathogen]] microorganisms to use in public health, outbreaks detection, agriculture and environment;<ref>{{Cite web |title=100K Pathogen Genome Project – Genomes for Public Health & Food Safety |url=https://100kgenomes.org/ |access-date=2022-08-18 |language=en-US}}</ref> the [[Earth BioGenome Project]], which started in 2018 and aims to sequence and catalog the genomes of all the eukaryotic organisms on Earth to promote biodiversity conservation projects. Inside this big-science project there are up to 50 smaller-scale affiliated projects such as the [[Africa BioGenome Project]] or the [[1000 Fungal Genomes Project]].<ref>{{cite journal | vauthors = Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, Durbin R, Edwards SV, Forest F, Gilbert MT, Goldstein MM, Grigoriev IV, Hackett KJ, Haussler D, Jarvis ED, Johnson WE, Patrinos A, Richards S, Castilla-Rubio JC, van Sluys MA, Soltis PS, Xu X, Yang H, Zhang G | display-authors = 6 | title = Earth BioGenome Project: Sequencing life for the future of life | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 115 | issue = 17 | pages = 4325–4333 | date = April 2018 | pmid = 29686065 | pmc = 5924910 | doi = 10.1073/pnas.1720115115 }}</ref><ref>{{Cite web |title=African BioGenome Project – Genomics in the service of conservation and improvement of African biological diversity |url=https://africanbiogenome.org/ |access-date=2022-08-18 |language=en-US}}</ref><ref>{{Cite web |title=1000 Fungal Genomes Project |url=https://mycocosm.jgi.doe.gov/mycocosm/home/1000-fungal-genomes |access-date=2022-08-18 |website=mycocosm.jgi.doe.gov}}</ref> |
||
Revision as of 13:02, 18 August 2022

A reference genome (also known as a reference assembly) is a digital nucleic acid sequence database, assembled by scientists as a representative example of the set of genes in one idealized individual organism of a species. As they are assembled from the sequencing of DNA from a number of individual donors, reference genomes do not accurately represent the set of genes of any single individual organism. Instead a reference provides a haploid mosaic of different DNA sequences from each donor. For example, the most recent human reference genome (assembly GRCh38/hg38) is derived from >60 genomic clone libraries. [1] There are reference genomes for multiple species of viruses, bacteria, fungus, plants, and animals. Reference genomes are typically used as a guide on which new genomes are built, enabling them to be assembled much more quickly and cheaply than the initial Human Genome Project. Reference genomes can be accessed online at several locations, using dedicated browsers such as Ensembl or UCSC Genome Browser.[2]
Properties of reference genomes
Measures of length
The length of a genome can be measured in multiple different ways.
A simple way to measure genome length is to count the number of base pairs in the assembly.[3]
The golden path is an alternative measure of length that omits redundant regions such as haplotypes and pseudoautosomal regions.[4][5] It is usually constructed by layering sequencing information over a physical map to combine scaffold information. It is a 'best estimate' of what the genome will look like and typically includes gaps, making it longer than the typical base pair assembly.[6]
Mammalian genomes
The human and mouse reference genomes are maintained and improved by the Genome Reference Consortium (GRC), a group of fewer than 20 scientists from a number of genome research institutes, including the European Bioinformatics Institute, the National Center for Biotechnology Information, the Sanger Institute and McDonnell Genome Institute at Washington University in St. Louis. GRC continues to improve reference genomes by building new alignments that contain fewer gaps, and fixing misrepresentations in the sequence.
Human reference genome
The original human reference genome was derived from thirteen anonymous volunteers from Buffalo, New York. Donors were recruited by advertisement in The Buffalo News, on Sunday, March 23, 1997. The first ten male and ten female volunteers were invited to make an appointment with the project's genetic counselors and donate blood from which DNA was extracted. As a result of how the DNA samples were processed, about 80 percent of the reference genome came from eight people and one male, designated RP11, accounts for 66 percent of the total. The ABO blood group system differs among humans, but the human reference genome contains only an O allele, although the others are annotated.[7][8][9][10][11]

As the cost of DNA sequencing falls, and new full genome sequencing technologies emerge, more genome sequences continue to be generated. In several cases people such as James D. Watson had their genome assembled using massive parallel DNA sequencing.[12][13] Comparison between the reference (assembly NCBI36/hg18) and Watson's genome revealed 3.3 million single nucleotide polymorphism differences, while about 1.4 percent of his DNA could not be matched to the reference genome at all.[11][12] For regions where there is known to be large-scale variation, sets of alternate loci are assembled alongside the reference locus.
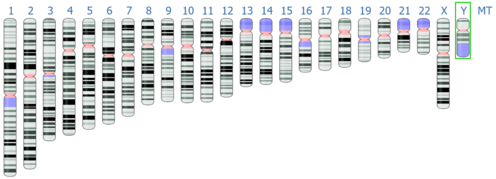
The latest patch of the human reference genome released by the Genome Reference Consortium was GRCh38 in 2017.[15] This build only has 120 gaps across all the assembly, which implies a great improvement in comparison with previous genome assemblies; the first version had roughly 150,000 gaps.[8] It presents gaps mostly in areas concerning telomeres, centromeres and long repetitive sequences, being the biggest gap along the long arm of the Y chromosome, a region of ~30 Mb length (~52% of the Y chromosome length).[16] The number of genomic clone libraries contributing to the reference has increased steadily to >60 along the years, although individual RP11 still accounts for 70% of the genome. [1] Genomic analysis of this anonymous male suggests that he is of African-European ancestry. [1] In 2022, the Telomere-to-Telomere (T2T) Consortium[17] published the first completely assembled reference genome (version T2T-CHM13), without any gaps in the assembly.[18][19] On the other hand, according to the GRC website, their next assembly release for the human genome (version GRCh39) is currently "indefinitely postponed".[20]
Recent genome assemblies are as follows:[21]
| Release name | Date of release | Equivalent UCSC version |
|---|---|---|
| GRCh39 | Indefinitely postponed[20] | - |
| T2T-CHM13 | January 2022 | - |
| GRCh38 | Dec 2013 | hg38 |
| GRCh37 | Feb 2009 | hg19 |
| NCBI Build 36.1 | Mar 2006 | hg18 |
| NCBI Build 35 | May 2004 | hg17 |
| NCBI Build 34 | Jul 2003 | hg16 |
Limitations
For much of a genome, the reference provides a good approximation of the DNA of any single individual. But in regions with high allelic diversity, such as the major histocompatibility complex in humans and the major urinary proteins of mice, the reference genome may differ significantly from other individuals.[22][23][24] Due to the fact that the reference genome is a "single" distinct sequence, which gives its utility as an index or locator of genomic features, there are limitations in terms of how faithfully it represents the human genome and its variability. On the other hand, most of the samples used for reference genomes sequencing come from people of european ancestry, being these human populations the best characterized and studied at the expense of non-european populations. In 2010, it was found that, by de novo assembling genomes from african and asian populations with the NCBI reference genome (version NCBI36), these genomes had ~5Mb sequences that did not align against any region of the reference genome.[25]
The 1000 Genomes Project is creating a database to provide information about the variations in genomes across human populations, which the reference genome is not able to represent.[26]
Mouse reference genome
Recent mouse genome assemblies are as follows:[21]
| Release name | Date of release | Equivalent UCSC version |
|---|---|---|
| GRCm39 | June 2020 | mm39 |
| GRCm38 | Dec 2011 | mm10 |
| NCBI Build 37 | Jul 2007 | mm9 |
| NCBI Build 36 | Feb 2006 | mm8 |
| NCBI Build 35 | Aug 2005 | mm7 |
| NCBI Build 34 | Mar 2005 | mm6 |
Other genomes
Since the Human Genome Project was finished, multiple international projects have started, focused on assembling reference genomes for many organisms. Model organisms (e.g., zebrafish (Danio rerio), chicken (Gallus gallus), Escherichia coli etc.) are of special interest to the scientific community, as well as, for example, endangered species (e.g., Asian arowana (Scleropages formosus) or the American bison (Bison bison)). As of August 2022, the NCBI database supports 71 886 partially or completely sequenced and assembled genomes from different species, such as 676 mammals, 590 birds and 865 fishes. Also noteworthy are the numbers of 1796 insects genomes, 3747 fungi, 1025 plants, 33 724 bacteria, 26 004 virus and 2040 archaea.[27] A lot of these species have annotation data associated with their reference genomes that can be publicly accessed and visualized in genome browsers such as Ensembl and UCSC Genome Browser.[28][29]
Some examples of these international projects are: the Chimpanzee Genome Project, carried out between 2005 and 2013 jointly by the Broad Institute and the McDonnell Genome Institute of Washington University in St. Louis, which generated the first reference genomes for 4 subspecies of Pan troglodytes;[30][31] the 100K Pathogen Genome Project, which started in 2012 with the main goal of creating a database of reference genomes for 100 000 pathogen microorganisms to use in public health, outbreaks detection, agriculture and environment;[32] the Earth BioGenome Project, which started in 2018 and aims to sequence and catalog the genomes of all the eukaryotic organisms on Earth to promote biodiversity conservation projects. Inside this big-science project there are up to 50 smaller-scale affiliated projects such as the Africa BioGenome Project or the 1000 Fungal Genomes Project.[33][34][35]
References
- ^ a b c "How many individuals were sequenced for the human reference genome assembly?". Genome Reference Consortium. Retrieved 7 April 2022.
- ^ Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, et al. (January 2008). "Ensembl 2008". Nucleic Acids Research. 36 (Database issue): D707–D714. doi:10.1093/nar/gkm988. PMC 2238821. PMID 18000006.
- ^ "Help - Glossary - Homo sapiens - Ensembl genome browser 87". www.ensembl.org.
- ^ "Golden path length | VectorBase". www.vectorbase.org. Archived from the original on 2020-08-07. Retrieved 2016-12-12.
- ^ "Help - Glossary - Homo sapiens - Ensembl genome browser 87". www.ensembl.org.
- ^ "Whole assembly vs Golden path length in Ensembl? - SEQanswers". seqanswers.com. Retrieved 2016-12-12.
- ^ Scherer S (2008). A short guide to the human genome. CSHL Press. p. 135. ISBN 978-0-87969-791-4.
- ^ a b "E pluribus unum". Nature Methods. 7 (5): 331. May 2010. doi:10.1038/nmeth0510-331. PMID 20440876.
- ^ Ballouz S, Dobin A, Gillis JA (August 2019). "Is it time to change the reference genome?". Genome Biology. 20 (1): 159. doi:10.1186/s13059-019-1774-4. PMC 6688217. PMID 31399121.
- ^ Rosenfeld JA, Mason CE, Smith TM (11 July 2012). "Limitations of the human reference genome for personalized genomics". PloS One. 7 (7): e40294. Bibcode:2012PLoSO...740294R. doi:10.1371/journal.pone.0040294. PMC 3394790. PMID 22811759.
- ^ a b Wade N (May 31, 2007). "Genome of DNA Pioneer Is Deciphered". New York Times. Retrieved February 21, 2009.
- ^ a b Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. (April 2008). "The complete genome of an individual by massively parallel DNA sequencing". Nature. 452 (7189): 872–876. Bibcode:2008Natur.452..872W. doi:10.1038/nature06884. PMID 18421352.
- ^ The exception to this is J. Craig Venter whose DNA was sequenced and assembled using shotgun sequencing methods.
- ^ "Genome Data Viewer - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2022-08-18.
- ^ Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, et al. (May 2017). "Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly". Genome Research. 27 (5): 849–864. doi:10.1101/gr.213611.116. PMC 5411779. PMID 28396521.
- ^ "GRCh38.p14 - hg38 - Genome - Assembly - NCBI - Statistics Report". www.ncbi.nlm.nih.gov. Retrieved 2022-08-18.
- ^ "Telomere-to-Telomere". NHGRI. Retrieved 2022-08-16.
- ^ Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, et al. (April 2022). "The complete sequence of a human genome". Science. 376 (6588): 44–53. Bibcode:2022Sci...376...44N. doi:10.1126/science.abj6987. PMC 9186530. PMID 35357919. S2CID 247854936.
- ^ "T2T-CHM13v2.0 - Genome - Assembly - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2022-08-16.
- ^ a b "Genome Reference Consortium". www.ncbi.nlm.nih.gov. Retrieved 2022-08-18.
- ^ a b "UCSC Genome Bioinformatics: FAQ". genome.ucsc.edu. Retrieved 2016-08-18.
- ^ MHC Sequencing Consortium (October 1999). "Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium". Nature. 401 (6756): 921–923. Bibcode:1999Natur.401..921T. doi:10.1038/44853. PMID 10553908. S2CID 186243515.
- ^ Logan DW, Marton TF, Stowers L (September 2008). Vosshall LB (ed.). "Species specificity in major urinary proteins by parallel evolution". PloS One. 3 (9): e3280. Bibcode:2008PLoSO...3.3280L. doi:10.1371/journal.pone.0003280. PMC 2533699. PMID 18815613.
- ^ Hurst J, Beynon RJ, Roberts SC, Wyatt TD (October 2007). Urinary Lipocalins in Rodenta:is there a Generic Model?. Chemical Signals in Vertebrates 11. Springer New York. ISBN 978-0-387-73944-1.
- ^ Li, Ruiqiang; Li, Yingrui; Zheng, Hancheng; Luo, Ruibang; Zhu, Hongmei; Li, Qibin; Qian, Wubin; Ren, Yuanyuan; Tian, Geng; Li, Jinxiang; Zhou, Guangyu; Zhu, Xuan; Wu, Honglong; Qin, Junjie; Jin, Xin (January 2010). "Building the sequence map of the human pan-genome". Nature Biotechnology. 28 (1): 57–63. doi:10.1038/nbt.1596. ISSN 1546-1696.
- ^ "1000 Genomes | A Deep Catalog of Human Genetic Variation". Internationalgenome.org. Retrieved 2022-07-19.
- ^ "Genome List - Genome - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2022-08-18.
- ^ "Species List". uswest.ensembl.org. Retrieved 2022-08-18.
- ^ "GenArk: UCSC Genome Archive". hgdownload.soe.ucsc.edu. Retrieved 2022-08-18.
- ^ "Chimpanzee Genome Project". BCM-HGSC. 2016-03-04. Retrieved 2022-08-18.
- ^ Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, et al. (July 2013). "Great ape genetic diversity and population history". Nature. 499 (7459): 471–475. doi:10.1038/nature12228. PMC 3822165. PMID 23823723.
- ^ "100K Pathogen Genome Project – Genomes for Public Health & Food Safety". Retrieved 2022-08-18.
- ^ Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, et al. (April 2018). "Earth BioGenome Project: Sequencing life for the future of life". Proceedings of the National Academy of Sciences of the United States of America. 115 (17): 4325–4333. doi:10.1073/pnas.1720115115. PMC 5924910. PMID 29686065.
- ^ "African BioGenome Project – Genomics in the service of conservation and improvement of African biological diversity". Retrieved 2022-08-18.
- ^ "1000 Fungal Genomes Project". mycocosm.jgi.doe.gov. Retrieved 2022-08-18.
