Microfluorimetry: Difference between revisions
Srmendez15 (talk | contribs) |
Srmendez15 (talk | contribs) |
||
| Line 7: | Line 7: | ||
Microfluorimetry can be used as a method to distinguish different microorganisms from one another by analyzing and comparing the DNA content of each cell.<ref>{{cite journal |doi=10.1177/26.9.361883 |title=Quantation by flow microfluorometry of total cellular DNA in Acanthamoeba |year=1978 |last1=Coulson |first1=P. B. |last2=Tyndall |first2=R. |journal=Journal of Histochemistry & Cytochemistry |volume=26 |issue=9 |pages=713}}</ref> This same concept can also be applied to distinguish between cell types using a suitable fluorenscent dye which varies depending on purpose and is a critical technique in modern cell biology and genomics. <ref>{{cite journal|last=Lydon|first=MJ|coauthors=Keeler, KD; Thomas, DB|title=Vital DNA staining and cell sorting by flow microfluorometry.|journal=Journal of cellular physiology|date=1980 Feb|volume=102|issue=2|pages=175-81|pmid=6154714}}</ref> |
Microfluorimetry can be used as a method to distinguish different microorganisms from one another by analyzing and comparing the DNA content of each cell.<ref>{{cite journal |doi=10.1177/26.9.361883 |title=Quantation by flow microfluorometry of total cellular DNA in Acanthamoeba |year=1978 |last1=Coulson |first1=P. B. |last2=Tyndall |first2=R. |journal=Journal of Histochemistry & Cytochemistry |volume=26 |issue=9 |pages=713}}</ref> This same concept can also be applied to distinguish between cell types using a suitable fluorenscent dye which varies depending on purpose and is a critical technique in modern cell biology and genomics. <ref>{{cite journal|last=Lydon|first=MJ|coauthors=Keeler, KD; Thomas, DB|title=Vital DNA staining and cell sorting by flow microfluorometry.|journal=Journal of cellular physiology|date=1980 Feb|volume=102|issue=2|pages=175-81|pmid=6154714}}</ref> |
||
Another use of microfluorometry is [[flow cytometry]] which uses the emission of fluorochrome molecules and usually a laser as a light source to create data from particles and cells<ref>{{cite web|title=Flow Cytometry Basic|url=http://crl.berkeley.edu/flow_cytometry_basic.html|work=http://crl.berkeley.edu|accessdate=11/17/2012}}</ref>. It can be used to separate chromosomes at a vary high rate and used easily with next-gen sequencing. This technique can simply results by separating only the relevant chromosomes at a very fast rate. <ref>{{cite journal|last=Koshland|first=D|coauthors=Hieter, P|title=Visual assay for chromosome ploidy.|journal=Methods in enzymology|date=1987|volume=155|pages=351-72|pmid=PMC3431466|accessdate=11/17/2012}}</ref> For example, E. coli bacteriophages lambda and T4 were able to be |
Another use of microfluorometry is [[flow cytometry]] which uses the emission of fluorochrome molecules and usually a laser as a light source to create data from particles and cells<ref>{{cite web|title=Flow Cytometry Basic|url=http://crl.berkeley.edu/flow_cytometry_basic.html|work=http://crl.berkeley.edu|accessdate=11/17/2012}}</ref>. It can be used to separate chromosomes at a vary high rate and used easily with next-gen sequencing. This technique can simply results by separating only the relevant chromosomes at a very fast rate. <ref>{{cite journal|last=Koshland|first=D|coauthors=Hieter, P|title=Visual assay for chromosome ploidy.|journal=Methods in enzymology|date=1987|volume=155|pages=351-72|pmid=PMC3431466|accessdate=11/17/2012}}</ref> For example, E. coli bacteriophages lambda and T4 were able to be separated by flow cytometry which allowed for genomic analysis which was previously difficult.<ref>{{cite journal|last=Allen|first=LZ|coauthors=Ishoey, T; Novotny, MA; McLean, JS; Lasken, RS; Williamson, SJ|title=Single virus genomics: a new tool for virus discovery.|journal=PloS one|date=2011 Mar 23|volume=6|issue=3|pages=e17722|pmid=21436882}}</ref> |
||
=== Concept === |
=== Concept === |
||
Revision as of 04:01, 19 November 2012
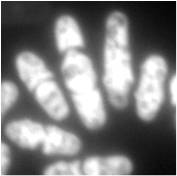
Microfluorimetry is a adaption of fluorimetry for studying the biochemical and biophysical properties of cells by using microscopy to image cell components tagged with fluorescent molecules. It is a type of microphotometry that gives a quantitative measure of the qualitative nature of fluorescent measurement and therefore, allows for definitive results that would have been previously indiscernible to the naked eye. [1]
Uses
Microfluorimetry has uses for many different fields including cell biology, microbiology, immunology, cell cycle analysis and "flow karyotyping" of cells.[2] In flow karotyping, isolated metaphase chromosomes are stained and measured in a flow microfluorometer. Fluorescent staining of chromosomes can also give distribution about the relative frequency of occurrence and the chromosomal DNA content of the measured chromosomes. This technique allows for karyotyping at higher speeds than with previous methods. [3] FMF can also be used to determine different populations of cells using fluorescent markers with small cell samples. The markers used for measurement in flow microfluorimetry are made up of fluorescent antigens or DNA binding agents.[2] It allows for the accurate measure of an antibody reacting with an antigen.[1]. Flow microfluorimetry is also used in pharmaceutical research to determine cell type, protein and DNA expression, cell cycle, and other properties of a cell during drug treatment. [4] Microfluorimetry can be used as a method to distinguish different microorganisms from one another by analyzing and comparing the DNA content of each cell.[5] This same concept can also be applied to distinguish between cell types using a suitable fluorenscent dye which varies depending on purpose and is a critical technique in modern cell biology and genomics. [6]
Another use of microfluorometry is flow cytometry which uses the emission of fluorochrome molecules and usually a laser as a light source to create data from particles and cells[7]. It can be used to separate chromosomes at a vary high rate and used easily with next-gen sequencing. This technique can simply results by separating only the relevant chromosomes at a very fast rate. [8] For example, E. coli bacteriophages lambda and T4 were able to be separated by flow cytometry which allowed for genomic analysis which was previously difficult.[9]
Concept
Microfluorimetry is building upon the established method of fluorimetric measurement. Using a dye that fluoresces in the presence of a target compound, fluorimetry can detect the presence of the compound by determining the presence and intensity of fluorescence. Differences in the intensity can be used to determine concentration of the compound. Additionally, if the dye undergoes a spectral shift then you can determine the absolute concentration of the target regardless of knowledge of the concentration of the dye. Fura-2 is an example of a fluorescent dye used to measure calcium. Microfluorimetry expands on fluorimetry by adding a microscopic component to measurements to allow analysis of single cells and other microscopic interests.[10]
Microfluorometer
A microfluorometer is a fluorescence spectrophotometer combined with a microscope, designed to measure fluorescence spectra of microscopic samples or areas or can be configured to measure the transmission and reflectance spectra of microscopic sample areas. It can either be a complete microfluorometer built exclusively for fluorescence microspectroscopy or the fluorescence spectrometer unit which attaches to the optical port of a microscope.[11] A microfluorometer can be used to estimate amounts and distributions of chemical components in individual cells or in chromosomes. In order to estimate the amount of chemical components, it's fluorescent intensity is measured by photoelectrical photometry while distribution is found by measuring the intensities of photos of negative chromosomes' metaphase plates.[12]. A microspectrophotometer can measure transmission, absorbance, reflectance and emission spectra then using built in algorithms a spectra is produced that can be compared against previous data in order to determine composition, concentration, etc.[13]
Limitations
There are many sources of error in the process but biological errors such as a inability to prepare homogenous samples are more likely to be a limitation than technical errors. [1]
See also
- Fluorescence spectroscopy, for a complete discussion of fluorescent measurement.
- Fluorometer, for a fuller description of the devices for fluorescent measurement.
References
- ^ a b c Goldman, Morris (1971). "Microfluorimetry in the Antigenic Analysis of Closely Related Microorganisms". Annals of the New York Academy of Sciences. 177: 439–45. doi:10.1111/j.1749-6632.1971.tb35073.x. PMID 4333753.
- ^ a b MacKenzie, NM; Pinder, AC (1986). "The application of flow microfluorimetry (FMF) to biomedical research and diagnosis: A review". Developments in biological standardization. 64: 181–93. PMID 3792646.
- ^ Gray, JW (1975 Aug). "High-speed quantitative karyotyping by flow microfluorometry". Clinical chemistry. 21 (9): 1258–62. PMID 1170959.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Siegel, EB (1984 Sep). "The use of flow microfluorometry for pharmaceutical testing". Regulatory toxicology and pharmacology : RTP. 4 (3): 287–304. PMID 6208577.
{{cite journal}}: Check date values in:|date=(help) - ^ Coulson, P. B.; Tyndall, R. (1978). "Quantation by flow microfluorometry of total cellular DNA in Acanthamoeba". Journal of Histochemistry & Cytochemistry. 26 (9): 713. doi:10.1177/26.9.361883.
- ^ Lydon, MJ (1980 Feb). "Vital DNA staining and cell sorting by flow microfluorometry". Journal of cellular physiology. 102 (2): 175–81. PMID 6154714.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ "Flow Cytometry Basic". http://crl.berkeley.edu. Retrieved 11/17/2012.
{{cite web}}: Check date values in:|accessdate=(help); External link in|work= - ^ Koshland, D (1987). "Visual assay for chromosome ploidy". Methods in enzymology. 155: 351–72. PMID PMC3431466.
{{cite journal}}:|access-date=requires|url=(help); Check|pmid=value (help); Check date values in:|accessdate=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Allen, LZ (2011 Mar 23). "Single virus genomics: a new tool for virus discovery". PloS one. 6 (3): e17722. PMID 21436882.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Smif, Pete. "Microfluorimetry". Retrieved 10/26/12.
{{cite web}}: Check date values in:|accessdate=(help) - ^ "Microspectrophotometer". microspectra.com. Craic Technologies. Retrieved 10/25/2012.
{{cite web}}: Check date values in:|accessdate=(help) - ^ Barskiĭ, IIa; Ioffe, VA; Kudriavtsev, BN; Papaian, GV; Rozanov, IuM (1983). "Microfluorimeter for chromosome study". Tsitologiia. 25 (6): 711–8. PMID 6351388.
- ^ "Microspectrophotometer". microspectra.com. Craic Technologies. Retrieved 10/25/2012.
{{cite web}}: Check date values in:|accessdate=(help)
External links
- http://microspectra.com/component/content/article/52-craictech/81-microfluorometer, Microfluorometer
- http://www.youtube.com/watch?v=_lmkEqTHj3Y, A video example of a microfluorimtery
