Cloning vector: Difference between revisions
| Line 10: | Line 10: | ||
All cloning vectors have features that allow a gene to be conveniently inserted into the vector or removed from it. This may be a [[multiple cloning site]] (MCS) which contains many unique [[restriction enzyme|restriction sites]]. The restriction sites in the MCS are first cleaved by restriction enzymes, and a [[PCR]]-amplified target gene, also digested with the same enzymes, is then ligated into the vectors using [[DNA ligase]]. The target DNA sequence can be inserted into the vector in a specific direction if so desired. The restriction sites may be further used for [[sub-cloning]] into another vector if necessary. |
All cloning vectors have features that allow a gene to be conveniently inserted into the vector or removed from it. This may be a [[multiple cloning site]] (MCS) which contains many unique [[restriction enzyme|restriction sites]]. The restriction sites in the MCS are first cleaved by restriction enzymes, and a [[PCR]]-amplified target gene, also digested with the same enzymes, is then ligated into the vectors using [[DNA ligase]]. The target DNA sequence can be inserted into the vector in a specific direction if so desired. The restriction sites may be further used for [[sub-cloning]] into another vector if necessary. |
||
Other cloning vectors may use [[topoisomerase]] instead of ligase and cloning may be done more rapidly without the need for restriction digest of the vector or insert. In this [[TOPO cloning]] method a linearized vector is activated by attaching topoisomerase I to its ends, and this "TOPO-activated" vector may then accept a PCR product by ligating both the 5' ends of the PCR product, releasing the topoisomerase and forming a circular vector in the process.<ref>{{cite web |url=http://www.invitrogen.com/site/us/en/home/brands/Product-Brand/topo/The-Technology-Behind-TOPO-Cloning.html |title=The Technology Behind TOPO® Cloning |work=Invitrogen }}</ref> Another method of cloning without the use of ligase is by [[Site-specific recombination|DNA recombination]], for example as used in the [[Gateway Technology|Gateway cloning system]]. |
Other cloning vectors may use [[topoisomerase]] instead of ligase and cloning may be done more rapidly without the need for restriction digest of the vector or insert. In this [[TOPO cloning]] method a linearized vector is activated by attaching topoisomerase I to its ends, and this "TOPO-activated" vector may then accept a PCR product by ligating both the 5' ends of the PCR product, releasing the topoisomerase and forming a circular vector in the process.<ref>{{cite web |url=http://www.invitrogen.com/site/us/en/home/brands/Product-Brand/topo/The-Technology-Behind-TOPO-Cloning.html |title=The Technology Behind TOPO® Cloning |work=Invitrogen }}</ref> Another method of cloning without the use of ligase is by [[Site-specific recombination|DNA recombination]], for example as used in the [[Gateway Technology|Gateway cloning system]].<ref>{{cite journal |title=Gateway cloning for protein expression|author= Esposito D, Garvey LA, Chakiath CS. |journal=Methods in Molelcular Biology |year= 2009 |volume=498|pages=31-54 |doi= 10.1007/978-1-59745-196-3_3 |pmid=18988017 }}</ref><ref>{{cite web |url=http://www.embl.de/pepcore/pepcore_services/cloning/cloning_methods/recombination/gateway/ |title=Cloning Methods - Recombination cloning systems |work=EMBL }}</ref> |
||
===Selectable marker=== |
===Selectable marker=== |
||
Revision as of 19:40, 2 February 2013
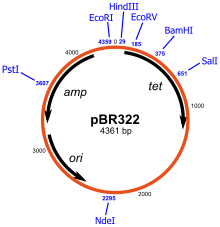
A cloning vector is a small piece of DNA, taken from a virus, a plasmid, or the cell of a higher organism, that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes.[1] The vector therefore contains features that allow for the convenient insertion or removal of DNA fragment in or out of the vector, for example by treating the vector and the foreign DNA with a restriction enzyme that creates the same overhang, then ligating the fragments together. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use.
There are many types of cloning vectors, but the most commonly-used ones are genetically engineered plasmids. Cloning is generally first performed using Escherichia coli, and cloning vectors in E. coli include plasmids, bacteriophages (such as phage λ), cosmids, and bacterial artificial chromosomes (BACs). Some DNA however cannot be stably maintained in E. coli, for example very large DNA fragment, and other organisms such as yeast may be used. Cloning vector in yeast include yeast artificial chromosomes (YACs).
Features of a cloning vector
All commonly-used cloning vectors in molecular biology have key features necessary for their function such as a suitable cloning site and selectable marker. Others may have additional features specific to their use. For reason of ease and convenience, cloning is often performed using E. coli, the cloning vectors used therefore often have elements necessary for their propagation and maintenance in E. coli such as a functional origin of replication (ori). The ColE1 origin of replication is found in many plasmids. Some cloning vectors also include elements that allow them to be maintained in another organism in addition to E. coli, and these vectors are called shuttle vector.
Cloning site
All cloning vectors have features that allow a gene to be conveniently inserted into the vector or removed from it. This may be a multiple cloning site (MCS) which contains many unique restriction sites. The restriction sites in the MCS are first cleaved by restriction enzymes, and a PCR-amplified target gene, also digested with the same enzymes, is then ligated into the vectors using DNA ligase. The target DNA sequence can be inserted into the vector in a specific direction if so desired. The restriction sites may be further used for sub-cloning into another vector if necessary.
Other cloning vectors may use topoisomerase instead of ligase and cloning may be done more rapidly without the need for restriction digest of the vector or insert. In this TOPO cloning method a linearized vector is activated by attaching topoisomerase I to its ends, and this "TOPO-activated" vector may then accept a PCR product by ligating both the 5' ends of the PCR product, releasing the topoisomerase and forming a circular vector in the process.[2] Another method of cloning without the use of ligase is by DNA recombination, for example as used in the Gateway cloning system.[3][4]
Selectable marker
A selectable marker is carried by the vector to allow the selection of positively transformed cells. Antibiotic resistance is often used as marker, an example is the beta-lactamase gene which confers resistance to the penicillin group of beta-lactam antibiotics like ampicillin.
Elements for expression
A cloning vector need not contain suitable elements for the expression of a cloned target gene, many however do, and may then work as an expression vector. The target DNA may be inserted into a site that is under the control of a particular promoter necessary for the expression of the target gene in the chosen host. Where the promoter is present, the expression of the gene is preferably tightly controlled. Some commonly used promoters are T7 promoters, lac promoters and cauliflower mosaic virus's 35s promoter (for plant vectors). The presence of a promoter is necessary when screening techniques such as blue-white selection are used.
Cloning vectors without promoter and ribosomal binding site (RBS) for the cloned DNA sequence are sometimes used, for example when cloning genes whose products are toxic to E. coli cells. Promoter and RBS for the cloned DNA sequence are also unnecessary when making a cDNA library of clones since the cloned genes are normally subcloned into a more appropriate expression vector.
Others
Some other possible features present in cloning vectors are: vir genes for plant transformation, integrase sites for chromosomal insertion, lacZα fragment for α complementation and blue-white selection, and/or reporter genes in frame with and flanking the MCS to facilitate the production of fusion proteins. Examples of a fusion partner are Green fluorescent protein (GFP) or glutathione S-transferase.
Screening: example of the blue/white selection

Many general purpose vectors such as pUC19 usually include a system for detecting the presence of a cloned DNA fragment, based on the loss of an easily scored phenotype. The most widely used is the gene coding for E. coli β-galactosidase, whose activity can easily be detected by the ability of the enzyme it encodes to hydrolyze the soluble, colourless substrate X-gal (5-bromo-4-chloro-3-indolyl-beta-d-galactoside) into an insoluble, blue product (5,5'-dibromo-4,4'-dichloro indigo). Cloning a fragment of DNA within the vector-based lacZα sequence of the β-galactosidase prevents the production of an active enzyme. If X-gal is included in the selective agar plates, transformant colonies are generally blue in the case of a vector with no inserted DNA and white in the case of a vector containing a fragment of cloned DNA.
See also
References
- ^ "Definition of cloning vector". Genome Dictionary. Retrieved 2012-10-18.
- ^ "The Technology Behind TOPO® Cloning". Invitrogen.
- ^ Esposito D, Garvey LA, Chakiath CS. (2009). "Gateway cloning for protein expression". Methods in Molelcular Biology. 498: 31–54. doi:10.1007/978-1-59745-196-3_3. PMID 18988017.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Cloning Methods - Recombination cloning systems". EMBL.
B. R. Glick and J. J. Pasternak (2005). Molecular Biotechnology Principles and Applications of Recombinant DNA. 3rd ed. ASM Press Washington, D. C.
