Phage typing: Difference between revisions
added a citation |
copied from sandbox https://en.wikipedia.org/wiki/User:Cafe1919/Phage_typing?veaction=edit&preload=Template:Dashboard.wikiedu.org_draft_template |
||
| Line 1: | Line 1: | ||
[[File:LambdaPlaques.jpg|right|thumb|A culture of bacteria infected by bacteriophages, the "holes" are areas where the bacteria have been killed by the virus. The culture is 10cm in diameter.]] |
[[File:LambdaPlaques.jpg|right|thumb|A culture of bacteria infected by bacteriophages, the "holes" are areas where the bacteria have been killed by the virus. The culture is 10cm in diameter.]] |
||
'''Phage typing''' is a method used for detecting single [[Strain (biology)|strains]] of [[bacteria]]. It is used to trace the source of outbreaks of infections.<ref name=" |
'''Phage typing''' is a method used for detecting single [[Strain (biology)|strains]] of [[bacteria]]. It is used to trace the source of outbreaks of infections.<ref name="pmid201223822">{{cite journal|author=Baggesen DL, Sørensen G, Nielsen EM, Wegener HC|year=2010|title=Phage typing of Salmonella Typhimurium - is it still a useful tool for surveillance and outbreak investigation?|url=http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19471|journal=[[Eurosurveillance]]|volume=15|issue=4|pages=19471|doi=|pmid=20122382}}</ref> The [[Virus|viruses]] that infect bacteria are called [[Bacteriophage|bacteriophages]] ("phages" for short) and some of these can only infect a single strain of bacteria. These phages are used to identify different strains of bacteria within a single species. The earliest recorded use of bacteriophages to identify bacteria dates back to 1925 when Sonnenschein used typhoid and paratyphoid phages to diagnose typhoid.<ref name=":6">{{Cite journal|last=Robertson|first=R. C.|last2=Yu|first2=H.|date=1936|title=The Vi antigen of B. typhosus|url=https://onlinelibrary.wiley.com/doi/abs/10.1002/path.1700430120|journal=The Journal of Pathology and Bacteriology|language=en|volume=43|issue=1|pages=191–196|doi=10.1002/path.1700430120|issn=1555-2039}}</ref> Phage typing capabilities were expanded with the discovery and adaptation of phages.<ref name=":1">{{Cite journal|last=Anderson|first=E. S.|last2=Ward|first2=Linda R.|last3=de Saxe|first3=Maureen J.|last4=de Sa|first4=J. D. H.|date=1977|title=Bacteriophage-Typing Designations of Salmonella typhimurium|url=https://www.jstor.org/stable/3861882|journal=The Journal of Hygiene|volume=78|issue=2|pages=297–300|issn=0022-1724}}</ref> |
||
== Principle == |
|||
| ⚫ | A culture of the strain is grown in the agar and dried. A grid is drawn on the base of the [[Petri dish]] to mark out different regions. Inoculation of each square of the grid is done by a different phage. The phage drops are allowed to dry and are incubated: The susceptible phage regions will show a circular clearing where the bacteria have been lysed, and this is used in differentiation.<ref name=" |
||
Phage typing is based on the specific binding of phages to antigens and receptors on the surface of bacteria and the resulting bacterial lysis or lack thereof.<ref name=":2">{{Cite journal|last=Felix|first=A.|last2=Callow|first2=B. R.|date=1943-07-31|title=Typing of Paratyphoid B Bacilli by Vi Bacteriophage|url=https://pubmed.ncbi.nlm.nih.gov/20784954/|journal=British Medical Journal|volume=2|issue=4308|pages=127–130|doi=10.1136/bmj.2.4308.127|issn=0007-1447|pmc=2284669|pmid=20784954}}</ref> The binding process is known as adsorption.<ref name=":0">{{Cite journal|last=Bertozzi Silva|first=Juliano|last2=Storms|first2=Zachary|last3=Sauvageau|first3=Dominic|date=2016-02-01|title=Host receptors for bacteriophage adsorption|url=https://doi.org/10.1093/femsle/fnw002|journal=FEMS Microbiology Letters|volume=363|issue=4|doi=10.1093/femsle/fnw002|issn=0378-1097}}</ref> Once a phage adsorbs to the surface of a bacteria, it may undergo either the lytic cycle or the lysogenic cycle.<ref name=":3">{{Cite journal|last=Kropinski|first=Andrew M|date=2006|title=Phage Therapy – Everything Old is New Again|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2095089/|journal=The Canadian Journal of Infectious Diseases & Medical Microbiology|volume=17|issue=5|pages=297–306|issn=1712-9532|pmc=2095089|pmid=18382643}}</ref> |
|||
Virulent phages enter the lytic pathway where they replicate and lyse the bacterial cell.<ref name=":4">{{Cite web|date=2018-07-14|title=21.2B: The Lytic and Lysogenic Cycles of Bacteriophages|url=https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Book%3A_General_Biology_(Boundless)/21%3A_Viruses/21.2%3A_Virus_Infections_and_Hosts/21.2B%3A_The_Lytic_and_Lysogenic_Cycles_of_Bacteriophages|access-date=2021-10-31|website=Biology LibreTexts|language=en}}</ref> Virulent phages can differentiate between different species of bacteria based on their specific lytic action.<ref>{{Cite journal|last=Koskella|first=Britt|last2=Meaden|first2=Sean|date=2013-03-11|title=Understanding bacteriophage specificity in natural microbial communities|url=https://pubmed.ncbi.nlm.nih.gov/23478639/|journal=Viruses|volume=5|issue=3|pages=806–823|doi=10.3390/v5030806|issn=1999-4915|pmc=3705297|pmid=23478639}}</ref> Lysis will only occur if the virulent phage adsorbs to the bacterial surface, configuring species specificity to phages.<ref name=":0" /> |
|||
Temperate phages enter the lysogenic cycle and do not immediately lyse the cell.<ref name=":4" /> The phage is instead integrated into the bacterial genome as a prophage during lysogenization, which protects the cell from being lysed by phages which are serologically identical or related.<ref name=":5">{{Cite journal|last=Boyd|first=J. S. K.|date=1956-07-21|title=Immunity of Lysogenic Bacteria|url=https://www.nature.com/articles/178141a0|journal=Nature|language=en|volume=178|issue=4525|pages=141–141|doi=10.1038/178141a0|issn=1476-4687}}</ref> Since it is incorporated into the genome, the prophage is also passed down to the bacteria's progenies.<ref name=":4" /> The bacterial strain carrying the prophage is known as a lysogenic strain.<ref name=":5" /> Lysogenization is strain-specific, so it allows for differentiation among different strains of bacteria within the same species.<ref>{{Cite web|title=Encyclopedia of Virology {{!}} ScienceDirect|url=https://www.sciencedirect.com/referencework/9780123744104/encyclopedia-of-virology|access-date=2021-11-15|website=www.sciencedirect.com}}</ref> The prophage may be chemically or physically induced to revert to the lytic pathway.<ref name=":3" /> |
|||
== Method == |
|||
| ⚫ | A culture of the strain is grown in the agar and dried. A grid is drawn on the base of the [[Petri dish]] to mark out different regions. Inoculation of each square of the grid is done by a different phage. The phage drops are allowed to dry and are incubated: The susceptible phage regions will show a circular clearing where the bacteria have been lysed, and this is used in differentiation.<ref name="pmid175822022">{{cite journal|vauthors=Turbadkar SD, Ghadge DP, Patil S, Chowdhary AS, Bharadwaj R|date=April 2007|title=Circulating phage type of Vibrio cholerae in Mumbai|url=http://www.ijmm.org/article.asp?issn=0255-0857;year=2007;volume=25;issue=2;spage=177;epage=178;aulast=Turbadkar|journal=Indian Journal of Medical Microbiology|volume=25|issue=2|pages=177–8|doi=10.4103/0255-0857.32738|pmid=17582202|doi-access=free}}</ref> The bacterial strain is characterized and epidemiologically identified by its susceptibility to different phages.<ref>{{Cite journal|last=Schofield|first=David A.|last2=Sharp|first2=Natasha J.|last3=Westwater|first3=Caroline|date=2012-04-01|title=Phage-based platforms for the clinical detection of human bacterial pathogens|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3442824/|journal=Bacteriophage|volume=2|issue=2|pages=105–283|doi=10.4161/bact.19274|issn=2159-7073|pmc=3442824|pmid=23050221}}</ref> |
||
== History == |
|||
The first reported use of bacteriophages to identify bacteria was in 1925 when Sonnenschein used typhoid and paratyphoid phages to diagnose typhoid.<ref>{{Citation|last=Brandis|first=Henning|title=Die Anwendung von Phagen in der bakteriologischen Diagnostik mit besonderer Berücksichtigung der Typisierung von Typhus- und Paratyphus B-Bakterien sowie Staphylokokken|date=1957|url=https://doi.org/10.1007/978-3-662-25832-3_3|work=Ergebnisse der Mikrobiologie, Immunitätsforschung und experimentellen Therapie: Fortsetzung der Ergebnisse der Hygiene, Bakteriologie, Immunitätsforschung und experimentellen Therapie|pages=96–159|editor-last=Kikuth|editor-first=W.|place=Berlin, Heidelberg|publisher=Springer|language=de|doi=10.1007/978-3-662-25832-3_3|isbn=978-3-662-25832-3|access-date=2021-11-15|editor2-last=Meyer|editor2-first=K. F.|editor3-last=Nauck|editor3-first=E. G.|editor4-last=Pappenheimer|editor4-first=A. M.}}</ref> In 1934, it was discovered that some strains of ''Salmonella typhi'' displayed Vi antigens on the surface.<ref name=":62">{{Cite journal|last=Robertson|first=R. C.|last2=Yu|first2=H.|date=1936|title=The Vi antigen of B. typhosus|url=https://onlinelibrary.wiley.com/doi/abs/10.1002/path.1700430120|journal=The Journal of Pathology and Bacteriology|language=en|volume=43|issue=1|pages=191–196|doi=10.1002/path.1700430120|issn=1555-2039}}</ref> This led to the isolation of Vi phages capable of lysing typhoid bacteria strains but only if they displayed the Vi antigen<ref>{{Cite journal|last=Scholtens|first=R. Th.|date=1937|title=Adsorption of vi Bacteriophages by Typhoid Bacilli and Paratyphoid C Strains|url=https://www.jstor.org/stable/3859853|journal=The Journal of Hygiene|volume=37|issue=2|pages=315–317|issn=0022-1724}}</ref> enabling the differentiation of typhoid species expressing the Vi antigen and those which do not. |
|||
In 1938, Craigie and Yen adapted Vi phages by selective propagation and used them at their critical test dilutions to differentiate 11 types of ''B. typhosus''.<ref>{{Cite journal|last=CRAIGIE|first=JAMES|last2=YEN|first2=CHUN HUI|date=1938|title=The Demonstration of Types of B. Typhosus by Means of Preparations of Type II Vi Phage: I. Principles and Technique|url=https://www.jstor.org/stable/41977832|journal=Canadian Public Health Journal|volume=29|issue=9|pages=448–463|issn=0319-2652}}</ref> In 1943, Felix and Callow extended the method to ''Salmonella paratyphi B''. in 1943 and differentiated 12 types with 11 phages.<ref name=":2" /> The International Committee for Enteric Phage Typing was established in 1947, and these phage typing methods were soon standardized.<ref>{{Cite web|title=International Federation for Enteric Phage Typing {{!}} UIA Yearbook Profile {{!}} Union of International Associations|url=https://uia.org/s/or/en/1100029572|access-date=2021-11-15|website=uia.org}}</ref> |
|||
Improvements to the specificity of phage typing schemes were made throughout the next few decades. In 1959, Callow improved her initial scheme to differentiate 34 types of Salmonella typhimurium with 29 phages.<ref name=":12">{{Cite journal|last=Anderson|first=E. S.|last2=Ward|first2=Linda R.|last3=de Saxe|first3=Maureen J.|last4=de Sa|first4=J. D. H.|date=1977|title=Bacteriophage-Typing Designations of Salmonella typhimurium|url=https://www.jstor.org/stable/3861882|journal=The Journal of Hygiene|volume=78|issue=2|pages=297–300|issn=0022-1724}}</ref> In 1977, this was extended to 207 types by Anderson at the Enteric Reference Laboratory in London.<ref name=":12" /> Since then, phage typing schemes have been developed for ''Salmonella typhi'', ''Salmonella paratyphi B.'', ''Salmonella typhimurium'', ''Shigella sonnei'', ''Staphylococcus aureus,'' and ''Escherichia coli'' to name a few.<ref>{{Cite journal|last=Anderson|first=E. S.|last2=Williams|first2=R. E.|date=May 1, 1956|title=Bacteriophage typing of enteric pathogens and staphylococci and its use in epidemiology|url=https://pubmed.ncbi.nlm.nih.gov/13332068/|journal=Journal of Clinical Pathology|volume=9|issue=2|pages=94–127|doi=10.1136/jcp.9.2.94|issn=0021-9746|pmc=1023924|pmid=13332068}}</ref><ref>{{Cite journal|last=Gershman|first=M.|last2=Merrill|first2=Cynthia E.|last3=Hunter|first3=Jacqueline|date=1981-12-01|title=Phage Typing Set for Differentiating Escherichia coli|url=https://www.sciencedirect.com/science/article/pii/S0022030281828627|journal=Journal of Dairy Science|language=en|volume=64|issue=12|pages=2392–2400|doi=10.3168/jds.S0022-0302(81)82862-7|issn=0022-0302}}</ref> |
|||
==References== |
==References== |
||
Revision as of 08:17, 15 November 2021
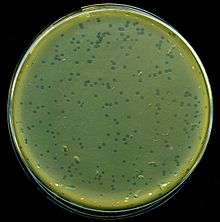
Phage typing is a method used for detecting single strains of bacteria. It is used to trace the source of outbreaks of infections.[1] The viruses that infect bacteria are called bacteriophages ("phages" for short) and some of these can only infect a single strain of bacteria. These phages are used to identify different strains of bacteria within a single species. The earliest recorded use of bacteriophages to identify bacteria dates back to 1925 when Sonnenschein used typhoid and paratyphoid phages to diagnose typhoid.[2] Phage typing capabilities were expanded with the discovery and adaptation of phages.[3]
Principle
Phage typing is based on the specific binding of phages to antigens and receptors on the surface of bacteria and the resulting bacterial lysis or lack thereof.[4] The binding process is known as adsorption.[5] Once a phage adsorbs to the surface of a bacteria, it may undergo either the lytic cycle or the lysogenic cycle.[6]
Virulent phages enter the lytic pathway where they replicate and lyse the bacterial cell.[7] Virulent phages can differentiate between different species of bacteria based on their specific lytic action.[8] Lysis will only occur if the virulent phage adsorbs to the bacterial surface, configuring species specificity to phages.[5]
Temperate phages enter the lysogenic cycle and do not immediately lyse the cell.[7] The phage is instead integrated into the bacterial genome as a prophage during lysogenization, which protects the cell from being lysed by phages which are serologically identical or related.[9] Since it is incorporated into the genome, the prophage is also passed down to the bacteria's progenies.[7] The bacterial strain carrying the prophage is known as a lysogenic strain.[9] Lysogenization is strain-specific, so it allows for differentiation among different strains of bacteria within the same species.[10] The prophage may be chemically or physically induced to revert to the lytic pathway.[6]
Method
A culture of the strain is grown in the agar and dried. A grid is drawn on the base of the Petri dish to mark out different regions. Inoculation of each square of the grid is done by a different phage. The phage drops are allowed to dry and are incubated: The susceptible phage regions will show a circular clearing where the bacteria have been lysed, and this is used in differentiation.[11] The bacterial strain is characterized and epidemiologically identified by its susceptibility to different phages.[12]
History
The first reported use of bacteriophages to identify bacteria was in 1925 when Sonnenschein used typhoid and paratyphoid phages to diagnose typhoid.[13] In 1934, it was discovered that some strains of Salmonella typhi displayed Vi antigens on the surface.[14] This led to the isolation of Vi phages capable of lysing typhoid bacteria strains but only if they displayed the Vi antigen[15] enabling the differentiation of typhoid species expressing the Vi antigen and those which do not.
In 1938, Craigie and Yen adapted Vi phages by selective propagation and used them at their critical test dilutions to differentiate 11 types of B. typhosus.[16] In 1943, Felix and Callow extended the method to Salmonella paratyphi B. in 1943 and differentiated 12 types with 11 phages.[4] The International Committee for Enteric Phage Typing was established in 1947, and these phage typing methods were soon standardized.[17]
Improvements to the specificity of phage typing schemes were made throughout the next few decades. In 1959, Callow improved her initial scheme to differentiate 34 types of Salmonella typhimurium with 29 phages.[18] In 1977, this was extended to 207 types by Anderson at the Enteric Reference Laboratory in London.[18] Since then, phage typing schemes have been developed for Salmonella typhi, Salmonella paratyphi B., Salmonella typhimurium, Shigella sonnei, Staphylococcus aureus, and Escherichia coli to name a few.[19][20]
References
- ^ Baggesen DL, Sørensen G, Nielsen EM, Wegener HC (2010). "Phage typing of Salmonella Typhimurium - is it still a useful tool for surveillance and outbreak investigation?". Eurosurveillance. 15 (4): 19471. PMID 20122382.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Robertson, R. C.; Yu, H. (1936). "The Vi antigen of B. typhosus". The Journal of Pathology and Bacteriology. 43 (1): 191–196. doi:10.1002/path.1700430120. ISSN 1555-2039.
- ^ Anderson, E. S.; Ward, Linda R.; de Saxe, Maureen J.; de Sa, J. D. H. (1977). "Bacteriophage-Typing Designations of Salmonella typhimurium". The Journal of Hygiene. 78 (2): 297–300. ISSN 0022-1724.
- ^ a b Felix, A.; Callow, B. R. (1943-07-31). "Typing of Paratyphoid B Bacilli by Vi Bacteriophage". British Medical Journal. 2 (4308): 127–130. doi:10.1136/bmj.2.4308.127. ISSN 0007-1447. PMC 2284669. PMID 20784954.
- ^ a b Bertozzi Silva, Juliano; Storms, Zachary; Sauvageau, Dominic (2016-02-01). "Host receptors for bacteriophage adsorption". FEMS Microbiology Letters. 363 (4). doi:10.1093/femsle/fnw002. ISSN 0378-1097.
- ^ a b Kropinski, Andrew M (2006). "Phage Therapy – Everything Old is New Again". The Canadian Journal of Infectious Diseases & Medical Microbiology. 17 (5): 297–306. ISSN 1712-9532. PMC 2095089. PMID 18382643.
- ^ a b c "21.2B: The Lytic and Lysogenic Cycles of Bacteriophages". Biology LibreTexts. 2018-07-14. Retrieved 2021-10-31.
- ^ Koskella, Britt; Meaden, Sean (2013-03-11). "Understanding bacteriophage specificity in natural microbial communities". Viruses. 5 (3): 806–823. doi:10.3390/v5030806. ISSN 1999-4915. PMC 3705297. PMID 23478639.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Boyd, J. S. K. (1956-07-21). "Immunity of Lysogenic Bacteria". Nature. 178 (4525): 141–141. doi:10.1038/178141a0. ISSN 1476-4687.
- ^ "Encyclopedia of Virology | ScienceDirect". www.sciencedirect.com. Retrieved 2021-11-15.
- ^ Turbadkar SD, Ghadge DP, Patil S, Chowdhary AS, Bharadwaj R (April 2007). "Circulating phage type of Vibrio cholerae in Mumbai". Indian Journal of Medical Microbiology. 25 (2): 177–8. doi:10.4103/0255-0857.32738. PMID 17582202.
- ^ Schofield, David A.; Sharp, Natasha J.; Westwater, Caroline (2012-04-01). "Phage-based platforms for the clinical detection of human bacterial pathogens". Bacteriophage. 2 (2): 105–283. doi:10.4161/bact.19274. ISSN 2159-7073. PMC 3442824. PMID 23050221.
- ^ Brandis, Henning (1957), Kikuth, W.; Meyer, K. F.; Nauck, E. G.; Pappenheimer, A. M. (eds.), "Die Anwendung von Phagen in der bakteriologischen Diagnostik mit besonderer Berücksichtigung der Typisierung von Typhus- und Paratyphus B-Bakterien sowie Staphylokokken", Ergebnisse der Mikrobiologie, Immunitätsforschung und experimentellen Therapie: Fortsetzung der Ergebnisse der Hygiene, Bakteriologie, Immunitätsforschung und experimentellen Therapie (in German), Berlin, Heidelberg: Springer, pp. 96–159, doi:10.1007/978-3-662-25832-3_3, ISBN 978-3-662-25832-3, retrieved 2021-11-15
- ^ Robertson, R. C.; Yu, H. (1936). "The Vi antigen of B. typhosus". The Journal of Pathology and Bacteriology. 43 (1): 191–196. doi:10.1002/path.1700430120. ISSN 1555-2039.
- ^ Scholtens, R. Th. (1937). "Adsorption of vi Bacteriophages by Typhoid Bacilli and Paratyphoid C Strains". The Journal of Hygiene. 37 (2): 315–317. ISSN 0022-1724.
- ^ CRAIGIE, JAMES; YEN, CHUN HUI (1938). "The Demonstration of Types of B. Typhosus by Means of Preparations of Type II Vi Phage: I. Principles and Technique". Canadian Public Health Journal. 29 (9): 448–463. ISSN 0319-2652.
- ^ "International Federation for Enteric Phage Typing | UIA Yearbook Profile | Union of International Associations". uia.org. Retrieved 2021-11-15.
- ^ a b Anderson, E. S.; Ward, Linda R.; de Saxe, Maureen J.; de Sa, J. D. H. (1977). "Bacteriophage-Typing Designations of Salmonella typhimurium". The Journal of Hygiene. 78 (2): 297–300. ISSN 0022-1724.
- ^ Anderson, E. S.; Williams, R. E. (May 1, 1956). "Bacteriophage typing of enteric pathogens and staphylococci and its use in epidemiology". Journal of Clinical Pathology. 9 (2): 94–127. doi:10.1136/jcp.9.2.94. ISSN 0021-9746. PMC 1023924. PMID 13332068.
- ^ Gershman, M.; Merrill, Cynthia E.; Hunter, Jacqueline (1981-12-01). "Phage Typing Set for Differentiating Escherichia coli". Journal of Dairy Science. 64 (12): 2392–2400. doi:10.3168/jds.S0022-0302(81)82862-7. ISSN 0022-0302.
[1][2][3][4][5][6][7][8][9][10][11]
- ^ Alamian, Dadar S, Solimani M, Behrozikhah S, Etemadi. 2019. A Case of Identity Confirmation of Brucella abortus S99 by Phage Typing and PCR Methods.
- ^ Cowley LA, Low AS, Pickard D, Boinett CJ, Dallman TJ, Day M, Perry N, Gally DL, Parkhill J, Jenkins C, et al. 2018. Transposon Insertion Sequencing Elucidates Novel Gene Involvement in Susceptibility and Resistance to Phages T4 and T7 in Escherichia coli.
- ^ Kaimal S, D’souza M, Sistla S, Parija SC. 2012. Phage typing in dermatitis cruris pustulosa et atrophicans: does staphylococcal carrier status have a role?
- ^ Lan R, Stevenson G, Donohoe K, Ward L, Reeves PR. 2007. Molecular markers with potential to replace phage typing for Salmonella enterica serovar typhimurium. J Microbiol Methods.
- ^ Lienemann T, Kyyhkynen A, Halkilahti J, Haukka K, Siitonen A. 2015. Characterization of Salmonella Typhimurium isolates from domestically acquired infections in Finland by phage typing, antimicrobial susceptibility testing, PFGE and MLVA. BMC Microbiol.
- ^ Prendergast DM, O’Grady D, Fanning S, Cormican M, Delappe N, Egan J, Mannion C, Fanning J, Gutierrez M. 2011. Application of multiple locus variable number of tandem repeat analysis (MLVA), phage typing and antimicrobial susceptibility testing to subtype Salmonella enterica serovar Typhimurium isolated from pig farms, pork slaughterhouses and meat producing plants in Ireland. Food Microbiol.
- ^ Schürch AC, van Soolingen D. 2012. DNA fingerprinting of Mycobacterium tuberculosis: From phage typing to whole-genome sequencing. Infect Genet Evol.
- ^ Szymczak P, Vogensen FK, Janzen T. 2019. Novel isolates of Streptococcus thermophilus bacteriophages from group 5093 identified with an improved multiplex PCR typing method. Int Dairy J.
- ^ Wiśniewska K, Szewczyk A, Piechowicz L, Bronk M, Samet A, Świeć K. 2012. The use of spa and phage typing for characterization of clinical isolates of methicillin-resistant Staphylococcus aureus in the University Clinical Center in Gdańsk, Poland. Folia Microbiol (Praha).
- ^ Xu D, Zhang J, Liu J, Xu J, Zhou H, Zhang L, Zhu J, Kan B. 2014. Outer Membrane Protein OmpW Is the Receptor for Typing Phage VP5 in the Vibrio cholerae O1 El Tor Biotype. J Virol.
- ^ Ziebell K, Chui L, King R, Johnson S, Boerlin P, Johnson RP. 2017. Subtyping of Canadian isolates of Salmonella Enteritidis using Multiple Locus Variable Number Tandem Repeat Analysis (MLVA) alone and in combination with Pulsed-Field Gel Electrophoresis (PFGE) and phage typing. J Microbiol Methods.
