Interactome: Difference between revisions
Alexbateman (talk | contribs) →References: Removed Stumpf ref that I've put inline |
Alexbateman (talk | contribs) Added inline ref for source of figure |
||
| Line 1: | Line 1: | ||
[[File:Network of how 100 of the 528 genes identified with significant differential expression relate to DISC1 and its core interactors.png|400px|right|thumb|Part of the [[DISC1]] interactome with genes represented by text in boxes and interactions noted by lines between the genes. From Hennah and Porteous, 2009.]] |
[[File:Network of how 100 of the 528 genes identified with significant differential expression relate to DISC1 and its core interactors.png|400px|right|thumb|Part of the [[DISC1]] interactome with genes represented by text in boxes and interactions noted by lines between the genes. From Hennah and Porteous, 2009.<ref name="pmid19300510">{{cite journal |author=Hennah W, Porteous D |title=The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes |journal=PLoS ONE |volume=4 |issue=3 |pages=e4906 |year=2009 |pmid=19300510 |pmc=2654149 |doi=10.1371/journal.pone.0004906 |url=}}</ref>]] |
||
In molecular biology an '''Interactome''' is defined as the whole set of molecular interactions in cells. Specifally it means physical interactions among molecules but can also mean indirect interactions among genes, i.e. [[genetic interactions]]. It is usually displayed as a [[directed graph]]. The word "interactome" was originally coined in 1999 by a group of French scientists headed by Bernard Jacq.<ref name="pmid9847149">{{cite journal |author=Sanchez C, Lachaize C, Janody F, ''et al.'' |title=Grasping at molecular interactions and genetic networks in Drosophila melanogaster using FlyNets, an Internet database |journal=Nucleic Acids Res. |volume=27 |issue=1 |pages=89–94 |year=1999 |month=January |pmid=9847149 |pmc=148104 |doi= |url=}}</ref> |
In molecular biology an '''Interactome''' is defined as the whole set of molecular interactions in cells. Specifally it means physical interactions among molecules but can also mean indirect interactions among genes, i.e. [[genetic interactions]]. It is usually displayed as a [[directed graph]]. The word "interactome" was originally coined in 1999 by a group of French scientists headed by Bernard Jacq.<ref name="pmid9847149">{{cite journal |author=Sanchez C, Lachaize C, Janody F, ''et al.'' |title=Grasping at molecular interactions and genetic networks in Drosophila melanogaster using FlyNets, an Internet database |journal=Nucleic Acids Res. |volume=27 |issue=1 |pages=89–94 |year=1999 |month=January |pmid=9847149 |pmc=148104 |doi= |url=}}</ref> |
||
Revision as of 13:18, 9 January 2012
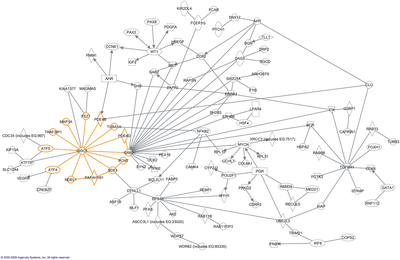
In molecular biology an Interactome is defined as the whole set of molecular interactions in cells. Specifally it means physical interactions among molecules but can also mean indirect interactions among genes, i.e. genetic interactions. It is usually displayed as a directed graph. The word "interactome" was originally coined in 1999 by a group of French scientists headed by Bernard Jacq.[2]
Molecular interaction networks
Molecular interactions can occur between molecules belonging to different biochemical families (proteins, nucleic acids, lipids, carbohydrates, etc.) and also within a given family. When spoken in terms of proteomics, interactome refers to protein–protein interaction network (PPI), or protein interaction network (PIN). Another extensively studied type of interactome is the protein–DNA interactome (network formed by transcription factors (and DNA or chromatin regulatory proteins) and their target genes.
Size of interactomes
It has been suggested that the size of an organism's interactome correlates better than genome size with the biological complexity of the organism.[3] Although protein–protein interaction maps containing several thousands of binary interactions are now available for several organisms, none of them is presently complete and the size of interactomes is still a matter of debate.
Genetic interaction networks
In 2010, the most "complete" gene interactome produced to date was compiled from 54 million two-gene comparisons to describe "the interaction profiles for ~75% of all genes in the Budding yeast," with 170,000 gene interactions.[4]
Although extremely important and useful, the interactome is still being developed and is not complete (as of October 2010[update]). There are various factors that have a role in protein interactions that have yet to be incorporated in the interactome. Many[who?] have termed the interactome as a whole as being fuzzy. The binding strength of the various proteins, microenvironmental factors, sensitivity to various procedures, and the physiological state of the cell all affect protein–protein interactions, yet are not accounted for in the interactome. Although the interactome is useful in some ways, it must be analyzed knowing that these factors exist and can affect the protein interactions.[5]
Methods of mapping the interactome
The study of the interactome is called interactomics. The basic unit of protein network is protein–protein interaction (PPI). Because the interactome considers the whole organism, there is a need to collect a massive amount of information.
Experimental methods have been devised to identify PPIs, such as affinity purification and the yeast two hybrid system (Y2H). The former is suited to identify a protein complex, while the latter is suited to explore the binary interactions among two proteins at a time. Both methods can be used in a high-throughput (HTP) fashion.
Using the experimental data as a starting point, the concept of homology transfer has been used to develop algorithms to map the interactome, including ones that produce detailed atomic models of protein protein complexes [6] as well as other protein–molecule interactions.[7]
There have been several efforts to map eukaryotic interactomes through HTP methods. As of 2006[update], yeast, fly, worm, and human HTP maps have been created. Recently, pathogen-host interactome (Hepatitis C Virus/Human (2008),[8] Epstein Barr virus/Human (2008), Influenza virus/Human (2009)) was also delineated through HTP to identify essential molecular components for pathoghens but also for the host to recognize pathogens and trigger efficient innate immune response.[9]
Using the interactome
Researchers have begun to use preliminary versions of the interactome to gain understanding about the biology and function of the molecules within them. For example, protein interaction networks have been used to produce improved protein functional annotations (or nannotations) for proteins with unknown functions.[10]
Interactome web servers
- Protinfo PPC predicts the atomic 3D structure of protein protein complexes.[11]
Interactome databases
- APID: Agile Protein Interaction DataAnalyzer – an interactive bioinformatic web-tool that integrates and unifies main known experimentally validated protein–protein interactions (Prieto and De Las Rivas, 2006).
- BioGRID database
- Bioverse database
- ConsensusPathDB includes functional interactions from 12 other databases
- Database of interacting proteins (DIP) (Xenarios et al. 2000).
- MIPS database
- PSIMAP database — the first protein structural interactome DB
- InterPare database — a structural protein interfaceome DB
- Biomolecular Interaction Network Database (BIND) (Bader et al. 2003)
- Online Predicted Human Interaction Database (OPHID) (Brown and Jurisica, 2005)
- Human Protein Reference Database (HPRD) (Peri S et al. 2003)
- HPID: Human Protein Interaction Database (Inha University, Korea)
- MINT: Molecular INTeraction database (Chatr-aryamontri et al. 2006)
- PINdb: Proteins Interacting in the Nucleus Database (Luc PV and Tempst P, 2004)
- IntAct: The Molecular Interaction Database
- VirHostNet — Virus-Host protein–protein interaction Networks knowledgebase
- Interactome Databases — interactome projects at CCSB.
- TRANSFAC, a database about transcription regulating eukaryotic protein-DNA interactions
See also
References
This article includes a list of references, related reading, or external links, but its sources remain unclear because it lacks inline citations. (March 2010) |
- ^ Hennah W, Porteous D (2009). "The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes". PLoS ONE. 4 (3): e4906. doi:10.1371/journal.pone.0004906. PMC 2654149. PMID 19300510.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Sanchez C, Lachaize C, Janody F; et al. (1999). "Grasping at molecular interactions and genetic networks in Drosophila melanogaster using FlyNets, an Internet database". Nucleic Acids Res. 27 (1): 89–94. PMC 148104. PMID 9847149.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Stumpf MP, Thorne T, de Silva E; et al. (2008). "Estimating the size of the human interactome". Proc. Natl. Acad. Sci. U.S.A. 105 (19): 6959–64. doi:10.1073/pnas.0708078105. PMC 2383957. PMID 18474861.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Costanzo M, Baryshnikova A, Bellay J; et al. (2010-01-22). "The genetic landscape of a cell". Science. 327 (5964): 425–431. doi:10.1126/science.1180823. PMID 20093466.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Welch GR (2008). "The Fuzzy Interactome" (PDF). Cell Press.
- ^ Kittichotirat W, Guerquin M, Bumgarner RE, Samudrala R. (2009.). "Protinfo PPC: A web server for atomic level prediction of protein complexes". Nucleic Acids Research. 37 (Web Server issue): W519–W525. doi:10.1093/nar/gkp306. PMC 2703994. PMID 19420059.
{{cite journal}}: Check date values in:|year=(help)CS1 maint: multiple names: authors list (link) - ^ McDermott J, Guerquin M, Frazier Z, Chang AN, Samudrala R. (2005). "BIOVERSE: Enhancements to the framework for structural, functional, and contextual annotations of proteins and proteomes". Nucleic Acids Research. 33 (Web Server issue): W324–W325. doi:10.1093/nar/gki401. PMC 1160162. PMID 15980482.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ de Chassey B, Navratil V, Tafforeau L; et al. (2008-11-04). "Hepatitis C virus infection protein network". Molecular Systems Biology. 4 (4:230): 230. doi:10.1038/msb.2008.66. PMC 2600670. PMID 18985028.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Navratil V, de Chassey B; et al. (2010-11-05). "Systems-level comparison of protein–protein interactions between viruses and the human type I interferon system network". Journal of Proteome Research. 9 (7): 3527–36. doi:10.1021/pr100326j. PMID 20459142.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ McDermott J, Bumgarner RE, Samudrala R. (2005). "Functional annotation from predicted protein interaction networks". Bioinformatics. 21 (15): 3217–3226. doi:10.1093/bioinformatics/bti514. PMID 15919725.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kittichotirat W, Guerquin M, Bumgarner R, Samudrala R. (2009). "Protinfo PPC: A web server for atomic level prediction of protein complexes". Nucleic Acids Research. 37: W519–W525.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
- De Las Rivas J, Fontanillo C (2010). "Protein–Protein Interactions Essentials: Key Concepts to Building and Analyzing Interactome Networks". PLoS Computational Biology. 6 (6): e1000807. doi:10.1371/journal.pcbi.1000807. PMC 2891586. PMID 20589078.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link). - Sanchez C, Lachaize C, Janody F, Bellon B, Röder L, Euzenat J, Rechenmann F, Jacq B (1999). "Grasping at molecular interactions and genetic networks in Drosophila melanogaster using FlyNets, an Internet database". Nucleic Acids Res. 27 (1): 89–94. doi:10.1093/nar/27.1.89. PMC 148104. PMID 9847149.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D (2000). "DIP: the database of interacting proteins". Nucleic Acids Res. 28 (1): 289–91. doi:10.1093/nar/28.1.289. PMC 102387. PMID 10592249.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Park J, Lappe M, Teichmann SA (2001). "Mapping protein family interactions: intramolecular and intermolecular protein family interaction repertoires in the PDB and yeast". J Mol Biol. 307 (3): 929–38. doi:10.1006/jmbi.2001.4526. PMID 11273711.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - Bader GD; et al. (2003). "BIND: The Biomolecular interaction Network Database". Nucleic Acids Res. 31 (1): 248–50. doi:10.1093/nar/gkg056. PMC 165503. PMID 12519993.
{{cite journal}}: Explicit use of et al. in:|author=(help) - Peri S., Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK; et al. (2003). "Development of human protein reference database as an initial platform for approaching systems biology in humans". Genome Res. 13 (10): 2363–71. doi:10.1101/gr.1680803. PMC 403728. PMID 14525934.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - Brown KR, Jurisica I (2005). "Online Predicted Human Interaction Database". Bioinformatics. 21 (9): 2076–82. doi:10.1093/bioinformatics/bti273. PMID 15657099.
- Navratil V. et al. (2009) VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res. 2009 Jan;37(Database issue):D661-8. Epub 2008 Nov 4.
External links
- PSIbase Database — a global interactome DB based on PSIMAP.
- Interactome.org — a dedicated interactome web site.
