RAB11B: Difference between revisions
m →Further reading: Journal cites, Added 1 doi to a journal cite using AWB (10365) |
Sarozpandey (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
{{PBB|geneid=9230}} |
{{PBB|geneid=9230}} |
||
'''Ras-related protein Rab-11B''' is a [[protein]] that in humans is encoded by the ''RAB11B'' [[gene]].<ref name="pmid7811277">{{cite journal | author = Zhu AX, Zhao Y, Flier JS | title = Molecular cloning of two small GTP-binding proteins from human skeletal muscle | journal = Biochem Biophys Res Commun | volume = 205 | issue = 3 | pages = 1875–82 |date=Feb 1995 | pmid = 7811277 | pmc = | doi = 10.1006/bbrc.1994.2889 }}</ref><ref name="entrez">{{cite web | title = Entrez Gene: RAB11B RAB11B, member RAS oncogene family| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=9230| accessdate = }}</ref> |
'''Ras-related protein Rab-11B''' is a [[protein]] that in humans is encoded by the ''RAB11B'' [[gene]].<ref name="pmid7811277">{{cite journal | author = Zhu AX, Zhao Y, Flier JS | title = Molecular cloning of two small GTP-binding proteins from human skeletal muscle | journal = Biochem Biophys Res Commun | volume = 205 | issue = 3 | pages = 1875–82 |date=Feb 1995 | pmid = 7811277 | pmc = | doi = 10.1006/bbrc.1994.2889 }}</ref><ref name="entrez">{{cite web | title = Entrez Gene: RAB11B RAB11B, member RAS oncogene family| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=9230| accessdate = }}</ref> |
||
==Introduction== |
|||
Rab (Ras-related in brain) proteins form the largest section of the Ras superfamily of small GTPases. The Rab family proteins regulate intracellular membrane trafficking processes including vesicle budding, tethering, and fusion. The isoforms Rab11a, Rab11b, and Rab11c/Rab25 constitute the Rab11 subfamily based on specific sequence motifs. {{cite journal |author=Bhartur SG, Calhoun BC, Woodrum J, ''et al.'' |title=Genomic structure of murine Rab11 family members |journal=Biochem. Biophys. Res. Commun. |volume=269 |issue=2 |pages=611–7 |year=2000 |month=March |pmid=10708602 |doi=10.1006/bbrc.2000.2334 |url=}} While RAB11A is located on chromosome 15 {{cite journal |author=Gromov PS, Celis JE, Hansen C, Tommerup N, Gromova I, Madsen P |title=Human rab11a: transcription, chromosome mapping and effect on the expression levels of host GTP-binding proteins |journal=FEBS Lett. |volume=429 |issue=3 |pages=359–64 |year=1998 |month=June |pmid=9662449 |doi= |url=}} and RAB11C on chromosome 1, RAB11B is placed on chromosome 19. Rab11 proteins are implicated in endocytosis and exocytosis. {{cite journal |author=Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J |title=Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network |journal=J. Cell Biol. |volume=151 |issue=6 |pages=1207–20 |year=2000 |month=December |pmid=11121436 |pmc=2190589 |doi= |url=}} Rab11b is reported as most abundantly expressed in brain, heart and testes. {{cite journal |author=Lai F, Stubbs L, Artzt K |title=Molecular analysis of mouse Rab11b: a new type of mammalian YPT/Rab protein |journal=Genomics |volume=22 |issue=3 |pages=610–6 |year=1994 |month=August |pmid=8001972 |doi=10.1006/geno.1994.1434 |url=}} Early studies with deletions of RAB11 homologs in Saccharomyces cerevisiae proved their importance in cell survival. {{cite journal |author=Benli M, Döring F, Robinson DG, Yang X, Gallwitz D |title=Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast |journal=EMBO J. |volume=15 |issue=23 |pages=6460–75 |year=1996 |month=December |pmid=8978673 |pmc=452471 |doi= |url=}} {{cite journal |author=Jedd G, Mulholland J, Segev N |title=Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment |journal=J. Cell Biol. |volume=137 |issue=3 |pages=563–80 |year=1997 |month=May |pmid=9151665 |pmc=2139891 |doi= |url=}} |
|||
Despite sharing high sequence homology, Rab11a and Rab11b appear to reside within distinct vesicle compartments. {{cite journal |author=Lapierre LA, Dorn MC, Zimmerman CF, Navarre J, Burnette JO, Goldenring JR |title=Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells |journal=Exp. Cell Res. |volume=290 |issue=2 |pages=322–31 |year=2003 |month=November |pmid=14567990 |doi= |url=}} Majority of Rab11b neither colocalize with transferrin receptor nor with the polymeric IgA receptor. This protein also exhibits a dependence on the microtubule cytoskeleton that is different from Rab11a. {{cite journal |author=Lapierre LA, Dorn MC, Zimmerman CF, Navarre J, Burnette JO, Goldenring JR |title=Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells |journal=Exp. Cell Res. |volume=290 |issue=2 |pages=322–31 |year=2003 |month=November |pmid=14567990 |doi= |url=}} High sequence diversity in the C-terminal hypervariable region is responsible for variable membrane targeting between these proteins. |
|||
==Functions== |
|||
Members of the Rab11 subfamily act in recycling of proteins from the endosomes to the plasma membrane, in transport of molecules from the trans-Golgi network to the plasma membrane and in phagocytosis. This subfamily also acts in polarized transport in epithelial cells. {{cite journal |author=Chen W, Feng Y, Chen D, Wandinger-Ness A |title=Rab11 is required for trans-golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor |journal=Mol. Biol. Cell |volume=9 |issue=11 |pages=3241–57 |year=1998 |month=November |pmid=9802909 |pmc=25617 |doi= |url=}}{{cite journal |author=Cox D, Lee DJ, Dale BM, Calafat J, Greenberg S |title=A Rab11-containing rapidly recycling compartment in macrophages that promotes phagocytosis |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=97 |issue=2 |pages=680–5 |year=2000 |month=January |pmid=10639139 |pmc=15390 |doi= |url=}}{{cite journal |author=Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI |title=The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform |journal=J. Struct. Biol. |volume=154 |issue=3 |pages=260–8 |year=2006 |month=June |pmid=16545962 |doi=10.1016/j.jsb.2006.01.007 |url=}}{{cite journal |author=Ullrich O, Reinsch S, Urbé S, Zerial M, Parton RG |title=Rab11 regulates recycling through the pericentriolar recycling endosome |journal=J. Cell Biol. |volume=135 |issue=4 |pages=913–24 |year=1996 |month=November |pmid=8922376 |pmc=2133374 |doi= |url=}}{{cite journal |author=Wang X, Kumar R, Navarre J, Casanova JE, Goldenring JR |title=Regulation of vesicle trafficking in madin-darby canine kidney cells by Rab11a and Rab25 |journal=J. Biol. Chem. |volume=275 |issue=37 |pages=29138–46 |year=2000 |month=September |pmid=10869360 |doi=10.1074/jbc.M004410200 |url=}} Whereas most studies refer to the Rab11a isoform, little is known about Rab11b so far. Rab11b localizes predominantly in the pericentriolar recycling compartment and serves as an important component of the vesicular machinery. {{cite journal |author=Schlierf B, Fey GH, Hauber J, Hocke GM, Rosorius O |title=Rab11b is essential for recycling of transferrin to the plasma membrane |journal=Exp. Cell Res. |volume=259 |issue=1 |pages=257–65 |year=2000 |month=August |pmid=10942597 |doi=10.1006/excr.2000.4947 |url=}} It is required for the transfer of internalized transferrin from the recycling compartment to the plasma membrane for which active Rab11b as well as GTP hydrolysis is necessary. {{cite journal |author=Schlierf B, Fey GH, Hauber J, Hocke GM, Rosorius O |title=Rab11b is essential for recycling of transferrin to the plasma membrane |journal=Exp. Cell Res. |volume=259 |issue=1 |pages=257–65 |year=2000 |month=August |pmid=10942597 |doi=10.1006/excr.2000.4947 |url=}} |
|||
==Clinical relevance== |
|||
Due to their crucial importance in vesicle transport and recycling, Rab11 proteins are linked to various non-pathogen or pathogen induced diseases. Most of the published data do not specify whether it is the a- or the b-isoform. |
|||
Rab11 proteins have been implicated in Alzheimer’s disease (14,15), Arthrogryposis-renal dysfunction-cholestasis (ARC) (16), Batten disease (17), and Charcot-Marie-Tooth Neuropathy Type 4C (CMT4C) (18). |
|||
Intracellular bacteria Chlamydia pneumoniae and Chlamydia trachomatis that replicate in membrane bound compartments hijack the trafficking machinery recruiting Rab GTPases to promote their replication within the host cell. Knock down of Rab11 decreased the formation of infectious particles (19–21). |
|||
Recent studies report a similar use of intracellular trafficking by Hantavirus and Influenza A virus. Replicated viruses benefit from Rab11 mediated recycling endosome pathway to exit the cell and infect surrounding tissue (22–25). |
|||
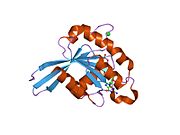
[[File:Rab11b-GNP-bind|framed|right|Cartoon model of active Rab11b-GppNP complex (PDB-ID: 2F9M {{cite journal |author=Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI |title=The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform |journal=J. Struct. Biol. |volume=154 |issue=3 |pages=260–8 |year=2006 |month=June |pmid=16545962 |doi=10.1016/j.jsb.2006.01.007 |url=}}). This model includes amino acids 8-188. The model shows classic Ras-like structure with a six stranded beta sheet core encircled by 5 major and one minor α-helix. GppNp and Mg2+ form polar bonds with the residues S20, K24, S25, N27, S40/S42/T43 (switch1), N124, K125, D127, S154, and with some parts of the backbone highlighted in olive green. Hydrogen-bonds are drawn as black dots. Atoms of GppNP, Mg2+, and sidechains are colored corresponding to their element. O = red, N = blue, P = orange, and Mg = green. The purple sequence represents the Rab-typical α310 helix region.]] |
|||
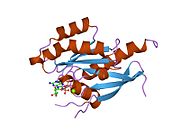
[[File:Alignment mod|framed|right|Alignment of inactive and active Rab11b (PDB-IDs: 2F9L/2F9M {{cite journal |author=Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI |title=The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform |journal=J. Struct. Biol. |volume=154 |issue=3 |pages=260–8 |year=2006 |month=June |pmid=16545962 |doi=10.1016/j.jsb.2006.01.007 |url=}}). The alignment shows slight conformational differences between the GDP- and the GppNP-bound form. Major divergences are displayed in red. The Rab11b-GDP structure shows the α310 helix in the switch 2 region whereas Rab11b-GppNP does not. Rab11b-GDP model lacks in the switch 1 section 39-41. These amino acids could not be modeled from the electron density map. Due the lack in switch 1 structure, the two states are not comparable from these models. Furthermore, the inactive state shows a truncated α-6 helix.]] |
|||
==Structure== |
|||
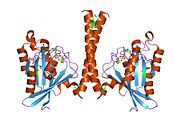
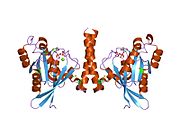
All Ras GTPases consist of a similar core structure and highly conserved P-loop, switch 1 and switch 2 regions. The Rab11b monomer exhibits a typical Ras-like, small GTPase fold with a six stranded β-sheet core (β1-β6) surrounded by five major α-helices (α1-α5) {{cite journal |author=Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI |title=The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform |journal=J. Struct. Biol. |volume=154 |issue=3 |pages=260–8 |year=2006 |month=June |pmid=16545962 |doi=10.1016/j.jsb.2006.01.007 |url=}} and one minor α-helix (α6). According to the sequence similarity to other Rab GTPases can be assumed that they show closely resembling characteristics in nucleotide binding and hydrolysis. However, Rab11 isoforms could differ in hydrolysis kinetics owing to the differences in conformation, since Rab11a and Rab11b do not show an α-helical switch 2 region like other Rab GTPases. Rab11b shares 90% amino acid identity to Rab11a. {{cite journal |author=Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI |title=The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform |journal=J. Struct. Biol. |volume=154 |issue=3 |pages=260–8 |year=2006 |month=June |pmid=16545962 |doi=10.1016/j.jsb.2006.01.007 |url=}} Kinetic experiments with Rab11a/b and Rab11-interacting proteins (FIPs) indicate that FIPs cannot differentiate between GTP-bound Rab11a and Rab11b in vitro (26). The major divergence reveals in the inactive state. While Pasqualato et al. crystallized inactive Rab11a as a dimer in the asymmetric unit, Scapin et al. observed single crystallographically independent monomers of both the GDP- and the GppNHp-bound Rab11b structures (10, 27). |
|||
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. --> |
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. --> |
||
Revision as of 14:21, 19 December 2014
Template:PBB Ras-related protein Rab-11B is a protein that in humans is encoded by the RAB11B gene.[1][2]
Introduction
Rab (Ras-related in brain) proteins form the largest section of the Ras superfamily of small GTPases. The Rab family proteins regulate intracellular membrane trafficking processes including vesicle budding, tethering, and fusion. The isoforms Rab11a, Rab11b, and Rab11c/Rab25 constitute the Rab11 subfamily based on specific sequence motifs. Bhartur SG, Calhoun BC, Woodrum J; et al. (2000). "Genomic structure of murine Rab11 family members". Biochem. Biophys. Res. Commun. 269 (2): 611–7. doi:10.1006/bbrc.2000.2334. PMID 10708602. {{cite journal}}: Explicit use of et al. in: |author= (help); Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) While RAB11A is located on chromosome 15 Gromov PS, Celis JE, Hansen C, Tommerup N, Gromova I, Madsen P (1998). "Human rab11a: transcription, chromosome mapping and effect on the expression levels of host GTP-binding proteins". FEBS Lett. 429 (3): 359–64. PMID 9662449. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) and RAB11C on chromosome 1, RAB11B is placed on chromosome 19. Rab11 proteins are implicated in endocytosis and exocytosis. Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J (2000). "Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network". J. Cell Biol. 151 (6): 1207–20. PMC 2190589. PMID 11121436. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) Rab11b is reported as most abundantly expressed in brain, heart and testes. Lai F, Stubbs L, Artzt K (1994). "Molecular analysis of mouse Rab11b: a new type of mammalian YPT/Rab protein". Genomics. 22 (3): 610–6. doi:10.1006/geno.1994.1434. PMID 8001972. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) Early studies with deletions of RAB11 homologs in Saccharomyces cerevisiae proved their importance in cell survival. Benli M, Döring F, Robinson DG, Yang X, Gallwitz D (1996). "Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast". EMBO J. 15 (23): 6460–75. PMC 452471. PMID 8978673. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) Jedd G, Mulholland J, Segev N (1997). "Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment". J. Cell Biol. 137 (3): 563–80. PMC 2139891. PMID 9151665. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link)
Despite sharing high sequence homology, Rab11a and Rab11b appear to reside within distinct vesicle compartments. Lapierre LA, Dorn MC, Zimmerman CF, Navarre J, Burnette JO, Goldenring JR (2003). "Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells". Exp. Cell Res. 290 (2): 322–31. PMID 14567990. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) Majority of Rab11b neither colocalize with transferrin receptor nor with the polymeric IgA receptor. This protein also exhibits a dependence on the microtubule cytoskeleton that is different from Rab11a. Lapierre LA, Dorn MC, Zimmerman CF, Navarre J, Burnette JO, Goldenring JR (2003). "Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells". Exp. Cell Res. 290 (2): 322–31. PMID 14567990. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) High sequence diversity in the C-terminal hypervariable region is responsible for variable membrane targeting between these proteins.
Functions
Members of the Rab11 subfamily act in recycling of proteins from the endosomes to the plasma membrane, in transport of molecules from the trans-Golgi network to the plasma membrane and in phagocytosis. This subfamily also acts in polarized transport in epithelial cells. Chen W, Feng Y, Chen D, Wandinger-Ness A (1998). "Rab11 is required for trans-golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor". Mol. Biol. Cell. 9 (11): 3241–57. PMC 25617. PMID 9802909. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link)Cox D, Lee DJ, Dale BM, Calafat J, Greenberg S (2000). "A Rab11-containing rapidly recycling compartment in macrophages that promotes phagocytosis". Proc. Natl. Acad. Sci. U.S.A. 97 (2): 680–5. PMC 15390. PMID 10639139. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link)Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI (2006). "The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform". J. Struct. Biol. 154 (3): 260–8. doi:10.1016/j.jsb.2006.01.007. PMID 16545962. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link)Ullrich O, Reinsch S, Urbé S, Zerial M, Parton RG (1996). "Rab11 regulates recycling through the pericentriolar recycling endosome". J. Cell Biol. 135 (4): 913–24. PMC 2133374. PMID 8922376. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link)Wang X, Kumar R, Navarre J, Casanova JE, Goldenring JR (2000). "Regulation of vesicle trafficking in madin-darby canine kidney cells by Rab11a and Rab25". J. Biol. Chem. 275 (37): 29138–46. doi:10.1074/jbc.M004410200. PMID 10869360. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) Whereas most studies refer to the Rab11a isoform, little is known about Rab11b so far. Rab11b localizes predominantly in the pericentriolar recycling compartment and serves as an important component of the vesicular machinery. Schlierf B, Fey GH, Hauber J, Hocke GM, Rosorius O (2000). "Rab11b is essential for recycling of transferrin to the plasma membrane". Exp. Cell Res. 259 (1): 257–65. doi:10.1006/excr.2000.4947. PMID 10942597. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) It is required for the transfer of internalized transferrin from the recycling compartment to the plasma membrane for which active Rab11b as well as GTP hydrolysis is necessary. Schlierf B, Fey GH, Hauber J, Hocke GM, Rosorius O (2000). "Rab11b is essential for recycling of transferrin to the plasma membrane". Exp. Cell Res. 259 (1): 257–65. doi:10.1006/excr.2000.4947. PMID 10942597. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link)
Clinical relevance
Due to their crucial importance in vesicle transport and recycling, Rab11 proteins are linked to various non-pathogen or pathogen induced diseases. Most of the published data do not specify whether it is the a- or the b-isoform. Rab11 proteins have been implicated in Alzheimer’s disease (14,15), Arthrogryposis-renal dysfunction-cholestasis (ARC) (16), Batten disease (17), and Charcot-Marie-Tooth Neuropathy Type 4C (CMT4C) (18). Intracellular bacteria Chlamydia pneumoniae and Chlamydia trachomatis that replicate in membrane bound compartments hijack the trafficking machinery recruiting Rab GTPases to promote their replication within the host cell. Knock down of Rab11 decreased the formation of infectious particles (19–21). Recent studies report a similar use of intracellular trafficking by Hantavirus and Influenza A virus. Replicated viruses benefit from Rab11 mediated recycling endosome pathway to exit the cell and infect surrounding tissue (22–25).
{{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link)). This model includes amino acids 8-188. The model shows classic Ras-like structure with a six stranded beta sheet core encircled by 5 major and one minor α-helix. GppNp and Mg2+ form polar bonds with the residues S20, K24, S25, N27, S40/S42/T43 (switch1), N124, K125, D127, S154, and with some parts of the backbone highlighted in olive green. Hydrogen-bonds are drawn as black dots. Atoms of GppNP, Mg2+, and sidechains are colored corresponding to their element. O = red, N = blue, P = orange, and Mg = green. The purple sequence represents the Rab-typical α310 helix region.{{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link)). The alignment shows slight conformational differences between the GDP- and the GppNP-bound form. Major divergences are displayed in red. The Rab11b-GDP structure shows the α310 helix in the switch 2 region whereas Rab11b-GppNP does not. Rab11b-GDP model lacks in the switch 1 section 39-41. These amino acids could not be modeled from the electron density map. Due the lack in switch 1 structure, the two states are not comparable from these models. Furthermore, the inactive state shows a truncated α-6 helix.Structure
All Ras GTPases consist of a similar core structure and highly conserved P-loop, switch 1 and switch 2 regions. The Rab11b monomer exhibits a typical Ras-like, small GTPase fold with a six stranded β-sheet core (β1-β6) surrounded by five major α-helices (α1-α5) Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI (2006). "The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform". J. Struct. Biol. 154 (3): 260–8. doi:10.1016/j.jsb.2006.01.007. PMID 16545962. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) and one minor α-helix (α6). According to the sequence similarity to other Rab GTPases can be assumed that they show closely resembling characteristics in nucleotide binding and hydrolysis. However, Rab11 isoforms could differ in hydrolysis kinetics owing to the differences in conformation, since Rab11a and Rab11b do not show an α-helical switch 2 region like other Rab GTPases. Rab11b shares 90% amino acid identity to Rab11a. Scapin SM, Carneiro FR, Alves AC, Medrano FJ, Guimarães BG, Zanchin NI (2006). "The crystal structure of the small GTPase Rab11b reveals critical differences relative to the Rab11a isoform". J. Struct. Biol. 154 (3): 260–8. doi:10.1016/j.jsb.2006.01.007. PMID 16545962. {{cite journal}}: Unknown parameter |month= ignored (help)CS1 maint: multiple names: authors list (link) Kinetic experiments with Rab11a/b and Rab11-interacting proteins (FIPs) indicate that FIPs cannot differentiate between GTP-bound Rab11a and Rab11b in vitro (26). The major divergence reveals in the inactive state. While Pasqualato et al. crystallized inactive Rab11a as a dimer in the asymmetric unit, Scapin et al. observed single crystallographically independent monomers of both the GDP- and the GppNHp-bound Rab11b structures (10, 27).
References
- ^ Zhu AX, Zhao Y, Flier JS (Feb 1995). "Molecular cloning of two small GTP-binding proteins from human skeletal muscle". Biochem Biophys Res Commun. 205 (3): 1875–82. doi:10.1006/bbrc.1994.2889. PMID 7811277.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Entrez Gene: RAB11B RAB11B, member RAS oncogene family".