Dendrogram: Difference between revisions
Adding a picture of dendrogram as a phylogenetic tree. |
A more compact hierarchical clustering picture. |
||
| Line 2: | Line 2: | ||
{{Refimprove|date=January 2017}} |
{{Refimprove|date=January 2017}} |
||
[[File:Iris dendrogram.png|thumb| [[Hierarchical clustering]] dendrogram of the [[Iris flower data set|Iris dataset]] (using [[R (programming language)|R]]). [https://cran.r-project.org/web/packages/dendextend/vignettes/Cluster_Analysis.html Source] ]] |
|||
[[File:UPGMA Dendrogram Hierarchical.svg|thumb| Dendrogram of a hierarchical clustering (UPGMA) with the height of the nodes.]] |
[[File:UPGMA Dendrogram Hierarchical.svg|thumb| Dendrogram of a hierarchical clustering (UPGMA) with the height of the nodes.]] |
||
[[File:Global-Diversity-of-Sponges-(Porifera)-pone.0035105.s008.tif|thumb|Dendrogram output for hierarchical clustering of marine provinces using presence / absence of sponge species.<ref name="VanSoest2012">{{Cite journal | author = Van Soest R, Boury-Esnault N, Vacelet J, Dohrmann M, Erpenbeck D, De Voogd N, Santodomingo N, Vanhoorne B, Kelly M, Hooper J | title = Global Diversity of Sponges (Porifera) | doi = 10.1371/journal.pone.0035105 | journal = PLOS ONE | year = 2012 | pmid = 22558119 | pmc = 3338747}} |
|||
</ref>]] |
|||
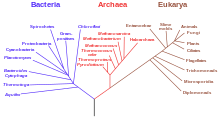
[[File:Phylogenetic tree.svg|thumb|A dendrogram of the [[Tree of Life]]. This phylogenetic tree is adapted from Woese et al. rRNA analysis.<ref name="Woese_1990">{{cite journal | last1 = Woese | first1 = Carl R.| authorlink1 = Carl Woese | last2 = [[Otto Kandler|Kandler]] | first2 = O | last3 = Wheelis | first3= M | title = Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya | url=http://www.pnas.org/content/87/12/4576.full.pdf | journal = Proc Natl Acad Sci USA | volume = 87 | issue = 12 | pages = 4576–4579 | year = 1990 | pmid = 2112744 | doi = 10.1073/pnas.87.12.4576 | pmc = 54159 | bibcode=1990PNAS...87.4576W}}</ref> The vertical line at bottom represents the [[last universal common ancestor]] (LUCA).]] |
[[File:Phylogenetic tree.svg|thumb|A dendrogram of the [[Tree of Life]]. This phylogenetic tree is adapted from Woese et al. rRNA analysis.<ref name="Woese_1990">{{cite journal | last1 = Woese | first1 = Carl R.| authorlink1 = Carl Woese | last2 = [[Otto Kandler|Kandler]] | first2 = O | last3 = Wheelis | first3= M | title = Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya | url=http://www.pnas.org/content/87/12/4576.full.pdf | journal = Proc Natl Acad Sci USA | volume = 87 | issue = 12 | pages = 4576–4579 | year = 1990 | pmid = 2112744 | doi = 10.1073/pnas.87.12.4576 | pmc = 54159 | bibcode=1990PNAS...87.4576W}}</ref> The vertical line at bottom represents the [[last universal common ancestor]] (LUCA).]] |
||
Revision as of 07:02, 23 October 2018
This article needs additional citations for verification. (January 2017) |


A dendrogram is a diagram representing a tree. This diagrammatic representation is frequently used in different contexts:
- in hierarchical clustering, it illustrates the arrangement of the clusters produced by the corresponding analyses.[3]
- in computational biology, it shows the clustering of genes or samples, sometimes in the margins of heatmaps.[4]
- in phylogenetics, it displays the evolutionary relationships among various biological taxa. In this case, the dendrogram is also called a phylogenetic tree.[5]
The name dendrogram derives from the two ancient greek words δένδρον (déndron), meaning "tree", and γράμμα (grámma), meaning "drawing, mathematical figure".[6][7]
Clustering example
For a clustering example, suppose that five taxa ( to ) have been clustered by UPGMA based on a matrix of genetic distances. The hierarchical clustering dendrogram would show a column of five nodes representing the initial data (here individual taxa), and the remaining nodes represent the clusters to which the data belong, with the arrows representing the distance (dissimilarity). The distance between merged clusters is monotone, increasing with the level of the merger: the height of each node in the plot is proportional to the value of the intergroup dissimilarity between its two daughters (the nodes on the right representing individual observations all plotted at zero height).
See also
- Cladogram
- Hierarchical clustering
- MEGA, a freeware for drawing dendrograms
- yEd, a freeware for drawing and automatically arranging dendrograms
- Distance matrices in phylogeny
References
- ^ Van Soest R, Boury-Esnault N, Vacelet J, Dohrmann M, Erpenbeck D, De Voogd N, Santodomingo N, Vanhoorne B, Kelly M, Hooper J (2012). "Global Diversity of Sponges (Porifera)". PLOS ONE. doi:10.1371/journal.pone.0035105. PMC 3338747. PMID 22558119.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Woese, Carl R.; Kandler, O; Wheelis, M (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya" (PDF). Proc Natl Acad Sci USA. 87 (12): 4576–4579. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- ^ Everitt, Brian (1998). Dictionary of Statistics. Cambridge, UK: Cambridge University Press. p. 96. ISBN 0-521-59346-8.
- ^ Wilkinson, Leland; Friendly, Michael (May 2009). "The History of the Cluster Heat Map". The American Statistician. 63 (2): 179–184. doi:10.1198/tas.2009.0033.
- ^ "Phylogenetic tree (biology)". Encyclopedia Britannica. Retrieved 2018-10-22.
- ^ Bailly, Anatole (1981-01-01). Abrégé du dictionnaire grec français. Paris: Hachette. ISBN 2010035283. OCLC 461974285.
- ^ Bailly, Anatole. "Greek-french dictionary online". www.tabularium.be. Retrieved October 20, 2018.
- Galili, T (2015). "dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering". Bioinformatics. 31: 3718–3720. doi:10.1093/bioinformatics/btv428. PMC 4817050. PMID 26209431.
External links
- Iris dendrogram - Example of using a dendrogram to visualize the 3 clusters from hierarchical clustering using the "complete" method vs the real species category (using R).


