HLA-A74
Appearance
| HLA-A74 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I, A cell surface antigen) | ||||||||||||||||
 HLA-A74 | ||||||||||||||||
| About | ||||||||||||||||
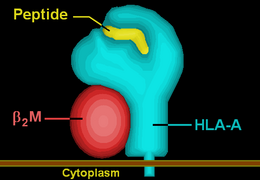
| Protein | transmembrane receptor/ligand | |||||||||||||||
| Structure | αβ heterodimer | |||||||||||||||
| Subunits | HLA-A*74--, β2-microglobulin | |||||||||||||||
| Subtypes | ||||||||||||||||
| ||||||||||||||||
| Rare alleles | ||||||||||||||||
| ||||||||||||||||
| Alleles link-out to IMGT/HLA database at EBI | ||||||||||||||||
HLA-A74 (A74) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α74 subset of HLA-A α-chains. For A74, the alpha "A" chain are encoded by the HLA-A*74 allele group and the β-chain are encoded by B2M locus.[1] A74 and A*74 are almost synonymous in meaning. A74 is a split antigen of the broad antigen serotype A19. A74 is a sister serotype of A29, A30, A31, A32, and A33.
A74 is more common in Subsaharan Africa. A74 is a rare HLA-A allele group.
Serotype
| A*74 | A74 | A19 | Sample |
| allele | % | % | size (N) |
| *7401 | 40 | 11 | 119 |
| *7403 | 17 | 0 | 29 |
A74 has a poor serotyping rate.
Disease associations
A significant association has been found between A74 and nasal polyposis.[3]
Allele distribution
| Study population | Freq. (in %)[4] |
|---|---|
| Colombia African Black | 11.6 |
| Kenya | 9.4 |
| Cameroon Beti | 8.3 |
| Guinea Bissau | 7.7 |
| Kenya Luo | 7.2 |
| Burkina Faso Mossi | 6.6 |
| South African Natal Zulu | 6.5 |
| Cape Verde Southeastern I… | 5.6 |
| Uganda Kampala | 5.2 |
| Cameroon Yaounde | 5.0 |
| Cameroon Bamileke | 4.5 |
| USA African America | 4.0 |
| Cameroon Sawa | 3.8 |
| South Africa Tswana | 3.7 |
| Mali Bandiagara | 3.6 |
| Iran Baloch | 2.8 |
| Zimbabwe Harare Shona | 2.7 |
| Zambia Lusaka | 2.3 |
| Senegal Niokholo Mandenka | 2.2 |
| Burkina Faso Rimaibe | 2.1 |
| Tunisia Ghannouch | 1.8 |
| Kenya Nandi | 1.7 |
| Brazil Parana Mulatto | 1.6 |
| Brazil | 1.5 |
| Morocco Nador Metalsa Cla… | 1.4 |
| Thailand Northeast | 1.2 |
| Venezuela Colonia Tovar | 1.2 |
| Brazil Belo Horizonte | 1.1 |
| Thailand | 1.1 |
| Tunisia Tunis | 1.1 |
| Turkey (1) | 1.1 |
| Allele frequencies presented, only | |
| Study population | Freq. (in %)[4] |
|---|---|
| Kenya Luo | 2.8 |
| Argentina Toba Rosario | 0.6 |
| Iran Baloch | 0.6 |
| Saudi Arabia Guraiat and … | 0.5 |
| Kenya | 0.3 |
| Madeira | 0.3 |
| Uganda Kampala | 0.3 |
| Allele frequencies presented, only | |
Sources
- Madrigal JA, Belich MP, Hildebrand WH, et al. (November 1992). "Distinctive HLA-A,B antigens of black populations formed by interallelic conversion" (PDF). J. Immunol. 149 (10): 3411–5. PMID 1431115.[permanent dead link]
References
- ^ Arce-Gomez B, Jones EA, Barnstable CJ, Solomon E, Bodmer WF (February 1978). "The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin". Tissue Antigens. 11 (2): 96–112. doi:10.1111/j.1399-0039.1978.tb01233.x. PMID 77067.
- ^ Allele Query Form IMGT/HLA - European Bioinformatics Institute
- ^ Luxenberger W, Posch U, Berghold A, Hofmann T, Lang-Loidolt D (2000). "HLA patterns in patients with nasal polyposis". European Archives of Oto-Rhino-Laryngology. 257 (3): 137–9. doi:10.1007/s004050050210. PMID 10839486.
- ^ a b Middleton, D.; Menchaca, L.; Rood, H.; Komerofsky, R. (2003). "New allele frequency database: http://www.allelefrequencies.net". Tissue Antigens. 61 (5): 403–407. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660.
