Molecular imaging
Molecular imaging originated from the field of radiopharmacology due to the need to better understand the fundamental molecular pathways inside organisms in a noninvasive manner.
Overview
Molecular Imaging is a new discipline that unites molecular biology and in vivo imaging. It enables the visualisation of the cellular function and the follow-up of the molecular process in living organisms without perturbing them. The multiple and numerous potentialities of this field are applicable to the diagnosis of diseases such as cancer, and neurological and cardiovascular diseases. This technique also contributes to improving the treatment of these disorders by optimizing the pre-clinical and clinical tests of new medication. They are also expected to have a major economic impact due to earlier and more precise diagnosis. Molecular and Functional Imaging has taken on a new direction since the description of the human genome. New paths in fundamental research, as well as in applied and industrial research, render the task of scientists more complex and increase the demands on them. Therefore, a tailor-made teaching program is in order.
Molecular imaging differs from traditional imaging in that probes known as biomarkers are used to help image particular targets or pathways. Biomarkers interact chemically with their surroundings and in turn alter the image according to molecular changes occurring within the area of interest. This process is markedly different from previous methods of imaging which primarily imaged differences in qualities such as density or water content. This ability to image fine molecular changes opens up an incredible number of exciting possibilities for medical application, including early detection and treatment of disease and basic pharmaceutical development. Furthermore, molecular imaging allows for quantitative tests, imparting a greater degree of objectivity to the study of these areas. One emerging technology is MALDI molecular imaging based on mass spectrometry.
Many areas of research are being conducted in the field of molecular imaging. Much research is currently centered around detecting what is known as a predisease state or molecular states that occur before typical symptoms of a disease are detected. Other important veins of research are the imaging of gene expression and the development of novel biomarkers. Organizations such as the SNM Molecular Imaging Center of Excellence (MICoE) have formed to support research in this field. In Europe, other "networks of excellence" such as DiMI (Diagnostics in Molecular Imaging) or EMIL (European Molecular Imaging Laboratories) work on this new science, integrating activities and research in the field. In this way, a European Master Programme "EMMI" is being set up to train a new generation of professionals in molecular imaging.
Recently the term "Molecular Imaging" has been applied to a variety of microscopy and nanoscopy techniques including live-cell microscopy, Total Internal Reflection Fluorescence (TIRF)-microscopy, STimulated Emission Depletion (STED)-nanoscopy and Atomic Force Microscopy (AFM) as here images of molecules are the readout.
Imaging modalities
There are many different modalities that can be used for noninvasive molecular imaging. Each have their different strengths and weaknesses and some are more adept at imaging multiple targets than others.
Magnetic resonance imaging (MRI)
MRI has the advantages of having very high spatial resolution and is very adept at morphological imaging and functional imaging. MRI does have several disadvantages though. First, MRI has a sensitivity of around 10-3 mol/L to 10-5 mol/L which compared to other types of imaging can be very limiting. This problem stems from the fact that the difference between atoms in the high energy state and the low energy state is very small. For example at 1.5 teslas the difference between high and low energy states is approximately 9 molecules per 2 million. Although with the use of small animal scanners much higher strength magnets can be used which can detect much lower concentrations than weaker magnets.
Optical imaging
There are a number of approaches used for optical imaging. The various methods depend upon fluorescence, bioluminescence, absorption or reflectance as the source of contrast.[1]
Optical imaging's most valuable attribute is that it and ultrasound do not have strong safety concerns like the other medical imaging modalities.
The downside of optical imaging is the lack of penetration depth, especially when working at visible wavelengths. Depth of penetration is related to the absorption and scattering of light, which is primarily a function of the wavelength of the excitation source. Light is absorbed by endogenous chromophores found in living tissue (e.g. hemoglobin, melanin, and lipids). In general, light absorption and scattering decreases with increasing wavelength. Below ~700 nm (e.g. visible wavelengths), these effects result in shallow penetration depths of only a few millimeters. Thus, in the visible region of the spectrum, only superficial assessment of tissue features is possible. Above 900 nm, water absorption can interfere with signal-to-background ratio. Because the absorption coefficient of tissue is considerably lower in the near infrared (NIR) region (700-900 nm), light can penetrate more deeply, to depths of several centimeters.[2]
Fluorescent probes and labels are an important tool for optical imaging. A number of near-infrared (NIR) fluorophores have been employed for in vivo imaging, including Kodak X-SIGHT Dyes and Conjugates, DyLight 750 and 800 Fluors, Cy 5.5 and 7 Fluors, Alexa Fluor 680 and 750 Dyes, IRDye 680 and 800 Fluors. Quantum dots, with their photostability and bright emissions, have generated a great deal of interest; however, their size precludes efficient clearance from the circulatory and renal systems while exhibiting long-term toxicity.
Several studies have demonstrated the use of infrared dye-labeled probes in optical imaging.
- In a comparison of gamma scintigraphy and NIR imaging, a cyclopentapeptide dual-labeled with 111indium and an NIR fluorophore was used to image αvβ3-integrin positive melanoma xenografts [3].
- An NIR fluorophore has been conjugated to epidermal growth factor (EGF) for imaging of tumor progression [4].
- An NIR fluorophore was compared to Cy5.5, suggesting that longer-wavelength dyes may produce more effective targeting agents for optical imaging [5].
- Pamidronate has been labeled with an NIR fluorophore and used as a bone imaging agent to detect osteoblastic activity in a living animal [6] .
- An NIR fluorophore-labeled GPI, a potent inhibitor of PSMA (prostate specific membrane antigen [7].
- Use of human serum albumin labeled with an NIR fluorophore as a tracking agent for mapping of sentinel lymph nodes [8].
- 2-deoxyglucose labeled with an NIR fluorophore [9].
Single photon emission computed tomography (SPECT)
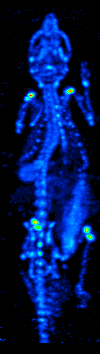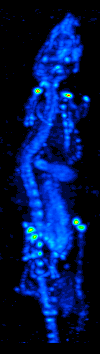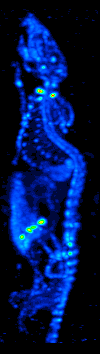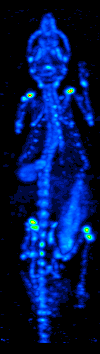
The main purpose of SPECT when used in brain imaging is to measure the regional cerebral blood flow (rCBF). The development of computed tomography in the 1970s allowed mapping of the distribution of the radioisotopes in the brain, and led to the technique now called SPECT.
The imaging agent used in SPECT emits gamma rays, as opposed to the positron emitters (such as 18F) used in PET. There are a range of radiotracers (such as 99mTc, 111In, 123I, 201Tl) that can be used, depending on the specific application.
Xenon (133Xe) gas is one such radiotracer. It has been shown to be valuable for diagnostic inhalation studies for the evaluation of pulmonary function; for imaging the lungs; and may also be used to assess rCBF. Detection of this gas occurs via a gamma camera—which is a scintillation detector consisting of a collimator, a NaI crystal, and a set of photomultiplier tubes.
By rotating the gamma camera around the head, a three dimensional image of the distribution of the radiotracer can be obtained by employing filtered back projection or other tomographic techniques. The radioisotopes used in SPECT have relatively long half lives (a few hours to a few days) making them easy to produce and relatively cheap. This represents the major advantage of SPECT as a brain imaging technique, since it is significantly cheaper than either PET or fMRI. However it lacks good spatial (i.e., where exactly the particle is) or temporal (i.e., did the contrast agent signal happen at this millisecond, or that millisecond) resolution. Additionally, due to the radioactivity of the contrast agent, there are safety aspects concerning the administration of radioisotopes to the subject, especially for serial studies.
Positron emission tomography (PET)
The theory behind PET is simple enough. First a molecule is tagged with a positron emitting isotope. These positrons annihilate with nearby electrons, emitting two 511,000 eV photons, directed 180 degrees apart in opposite directions. These photons are then detected by the scanner which can estimate the density of positron annihilations in a specific area. When enough interactions and annihilations have occurred, the density of the original molecule may be measured in that area. Typical isotopes include 11C, 13N, 15O, 18F, 64Cu, 62Cu, 124I, 76Br, 82Rb and 68Ga, with 18F being the most clinically utilized. One of the major disadvantages of PET is that most of the probes must be made with a cyclotron. Most of these probes also have a half life measured in hours, forcing the cyclotron to be on site. These factors can make PET prohibitively expensive. PET imaging does have many advantages though. First and foremost is its sensitivity: a typical PET scanner can detect between 10−11 mol/L to 10−12 mol/L concentrations.
Ultrasound
Explanation of how it works basics how it would be used in imaging multiple targets.
Probes and the imaging of molecular interactions
In order to image multiple targets you must first identify and develop the interactions of which you are trying to take advantage. Developing good probes is often difficult and is an area of intense research.
See also
- Predictive medicine
- Translational medicine
- Chemical imaging
- EMMI European Master in Molecular Imaging
References
- ^ Weissleder, R., Mahmood, U., Molecular Imaging, Radiology 2001; 219:316-333. | Download PDF
- ^ Olive D.M., Kovar, J.L., Simpson, M.A., Schutz-Geschwender, A., A systematic approach to the development of fluorescent contrast agents for optical imaging of mouse cancer models, Analytical Biochemistry 2007;(367), #1, 1-12. | Download PDF
- ^ Houston, J.P., Ke, S. Wang, W., Li, C., and Sevick-Muraca, E.M., J. Biomed. Optics 10, 054010 (2005)
- ^ Kovar, J.L., Johnson, M.A., Volcheck, W.M., Chen, J., and Simpson, M.A., Am. J. Pathol. 169, 1415 (2006). [1] | Download PDF]
- ^ Adams, K.E., Ke, S., Kwan, S., Liang, F., Fan, Z., Lu, Y., Barry, M.A., Mawad, M.E., and Sevick-Muraca, E.M., Journal of Biomedical Optics 12, 024017 (2007).
- ^ Zaheer, A., Lenkinski, R.E., Mahmood, A., Jones, A.G., Cantley, L.C., Frangioni, J.V., Nat. Biotechnol. 19, 1148 (2001).
- ^ Humblet, V., Lapidus, R., Williams, L.R., Tsukamoto, T., Rojas, C., Majer, P., Hin, B., Ohnishi, S., De Grand, A.M., Zaheer, A., Renze, J.T., Nakayama, A., Slusher, B.S., and Frangioni, J.V.. Molecular Imaging 4, 448 (2005).
- ^ Ohnishi, S., Lomnes, S.J., Laurence, R.G., Gogbashian, A., Mariani, G., and Frangioni, J.V., Molecular Imaging 4, 172 (2005).
- ^ Kovar, J., Sevick-Muraca, E., Simpson, M.A., and Olive, D.M., Abstract , World Molecular Imaging Conference, Nice, France.
Further reading
- Fuchs V.R.,Sox H.C. Jr., Health Affairs 2001: 20(5), 30–42
- Weissleder R, Mahmood U. Molecular imaging, Radiology 2001: 219:316-333
- Piwnica-worms D, Luker KE. Imaging Protein-protein interactions in whole cells and living animals. Ernst Schering Res Found Workshop. 2005;(49):35-41.
- Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light, Genes & Development 2003: 545-580
External links
- Society for Molecular Imaging
- Understanding Molecular Imaging and biomarker usage
- Annual Imaging Meeting - Annual conference on Imaging in clinical and pre-clinical imaging
- Molecular Imaging Research update from Reportergene
- Molecular Imaging Center
- Understanding Near-Infrared Imaging - Resource to better understand the benefits of Near-Infrared imaging.
- Society for Neuroscience -
- European Society for Molecular Imaging -
- European Institute for Biomedical Imaging Research -
