Bacterial conjugation
Bacterial conjugation is the transfer of genetic material between bacteria through direct cell to cell contact, or through a bridge-like connection between the two cells.[1] Discovered in 1946 by Joshua Lederberg and Edward Tatum,[2] conjugation is a mechanism of horizontal gene transfer—as are transformation and transduction—although these mechanisms do not involve cell-to-cell contact.[3]
Bacterial conjugation is often incorrectly regarded as the bacterial equivalent of sexual reproduction or mating. Loosely, and misleadingly, it can be considered to be a limited bacterial version of sex, since it involves some genetic exchange. In order to perform conjugation, one of the bacteria, the donor, must play host to a conjugative or mobilizable genetic element, most often a conjugative or mobilizable plasmid or transposon.[4][5] Most conjugative plasmids have systems ensuring that the recipient cell does not already contain a similar element.
The genetic information transferred is often beneficial to the recipient cell. Benefits may include antibiotic resistance, other xenobiotic tolerance, or the ability to utilize a new metabolite.[6] Such beneficial plasmids may be considered bacterial endosymbionts. Some conjugative elements may also be viewed as genetic parasites on the bacterium, and conjugation as a mechanism that was evolved by the mobile element to spread itself into new hosts.
Mechanism
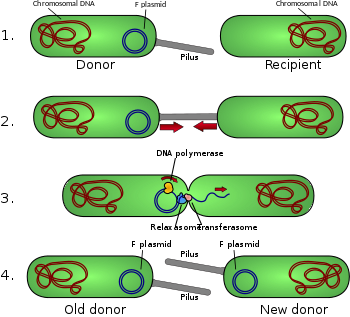
The prototype for conjugative plasmids is the F-plasmid, also called the F-factor.[1] The F-plasmid is an episome (a plasmid that can integrate itself into the bacterial chromosome by homologous recombination) of about 100 kb length. It carries its own origin of replication, the oriV, as well as an origin of transfer, or oriT.[4] There can only be one copy of the F-plasmid in a given bacterium, either free or integrated (two immediately before cell division). The host bacterium is called F-positive or F-plus (denoted F+). Strains that lack F plasmids are called F-negative or F-minus (F-).
Among other genetic information, the F-plasmid carries a tra and a trb locus, which together are about 33 kb long and consist of about 40 genes. The tra locus includes the pilin gene and regulatory genes, which together form pili on the cell surface, polymeric proteins that can attach themselves to the surface of F- bacteria and initiate the conjugation. Though there is some debate on the exact mechanism, the pili themselves do not seem to be the structures through which the actual exchange of DNA takes place. This has been shown through tests in which the pilus are allowed to make contact, then denatured by SDS detergent. DNA transformation still proceeds. Several proteins coded for in the tra or trb loci seem to open a channel between the bacteria. It is thought that the traD enzyme located at the base of the pilus is involved in DNA exchange, by iniating membrane fusion. [7].
When conjugation is initiated, via a mating signal, a relaxase enzyme creates a nick in one plasmid DNA strand at the origin of transfer, or oriT. The relaxase may work alone or in a complex of over a dozen proteins, known collectively as a relaxosome. In the F-plasmid system, the relaxase enzyme is called TraI and the relaxosome consists of TraI, TraY, TraM, and the integrated host factor, IHF. The transferred, or T-strand, is unwound from the duplex plasmid and transferred into the recipient bacterium in a 5'-terminus to 3'-terminus direction. The remaining strand is replicated, either independent of conjugative action (vegetative replication, beginning at the oriV) or in concert with conjugation (conjugative replication similar to the rolling circle replication of lambda phage). Conjugative replication may necessitate a second nick before successful transfer can occur. A recent report claims to have inhibited conjugation with chemicals that mimic an intermediate step of this second nicking event.[8]
If the F-plasmid becomes integrated into the host genome, donor chromosomal DNA may be transferred along with plasmid DNA.[3] The certain amount of chromosomal DNA that is transferred depends on how long the bacteria remain in contact; for common laboratory strains of E. coli the transfer of the entire bacterial chromosome takes about 100 minutes. The transferred DNA can be integrated into the recipient genome via homologous recombination.
A culture of cells containing non-integrated F plasmids usually contains a few that have accidentally become integrated, and these are responsible for those low-frequency chromosomal gene transfers which do occur in such cultures. Some strains of bacteria with an integrated F-plasmid can be isolated and grown in pure culture. Because such strains transfer chromosomal genes very efficiently, they are called Hfr (high frequency of recombination). The E. coli genome was originally mapped by interrupted mating experiments, in which various Hfr cells in the process of conjugation were sheared from recipients after less than 100 minutes (initially using a Waring blender) and investigating which genes were transferred.
Inter-kingdom transfer
The nitrogen fixing Rhizobia are an interesting case, wherein conjugative elements naturally engage in inter-kingdom conjugation[9]. Such elements as the Agrobacterium Ti or Ri plasmids contain elements that can transfer to plant cells. Transferred genes enter the plant cell nucleus and effectively transform the plant cells into factories for the production of opines, which the bacteria use as carbon and energy sources. Infected plant cells form crown gall or root tumors. The Ti and Ri plasmids are thus endosymbionts of the bacteria, which are in turn endosymbionts (or parasites) of the infected plant.
The Ti and Ri plasmids are themselves conjugative. Ti and Ri transfer between bacteria uses an independent system (the tra, or transfer, operon) from that for inter-kingdom transfer (the vir, or virulence, operon). Such transfer creates virulent strains from previously avirulent Agrobacteria.
Genetic Engineering Applications
Conjugation is a convenient means for transferring genetic material into a variety of targets. In the lab, successful transfer has been reported from bacteria into yeast[10], plants, mammalian cells[11][12], and isolated mammalian mitochondria[13]. Conjugation has advantages over some other forms of genetic transfer for engineering purposes, namely minimal disruption to the target's cellular envelope and the ability to transfer relatively large amounts of genetic material (see discussion of E. coli chromosome transfer, above). In plant engineering, Agrobacterium-like conjugation is complementary to other standard vehicles, such as tobacco mosaic virus (TMV). While TMV is capable of infecting many plant families, these are primarily herbaceous dicots. Agrobacterium-like conjugation is also primarily used for dicots, but monocot recipients are not uncommon.
See also
References
- ^ a b Holmes RK, Jobling MG (1996). Genetics: Conjugation. in: Baron's Medical Microbiology (Baron S et al., eds.) (4th ed.). Univ of Texas Medical Branch. ISBN 0-9631172-1-1.
- ^ Lederberg J, Tatum EL (1946). "Gene recombination in E. coli". Nature. 158: 558. doi:10.1038/158558a0.
- ^ a b Griffiths AJF; et al. (1999). An Introduction to genetic analysis (7th ed.). San Francisco: W.H. Freeman. ISBN 0-7167-3520-2.
{{cite book}}: Explicit use of et al. in:|author=(help) - ^ a b Ryan KJ, Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. pp. 60–4. ISBN 0838585299.
{{cite book}}:|author=has generic name (help) - ^ Russi; et al. (2008). "Molecular Machinery for DNA Translocation in Bacterial Conjugation". Plasmids: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-35-6.
{{cite book}}: Explicit use of et al. in:|author=(help); External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ Holmes RK, Jobling MG (1996). Genetics: Exchange of Genetic Information. in: Baron's Medical Microbiology (Baron S et al., eds.) (4th ed.). Univ of Texas Medical Branch. ISBN 0-9631172-1-1.
- ^ CITATION NEEDED BOOYAH
- ^ Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR (2007). "Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase". PNAS. 104: 12282–7. doi:10.1073/pnas.0702760104. PMC 1286216. PMID 17630285.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pan SQ, Jin S, Boulton MI, Hawes M, Gordon MP, Nester EW (1995). "An Agrobacterium virulence factor encoded by a Ti plasmid gene or a chromosomal gene is required for T-DNA transfer into plants". Mol. Microbiol. 17 (2): 259–69. doi:10.1111/j.1365-2958.1995.mmi_17020259.x. PMID 7494475.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Heinemann JA, Sprague GF (1989). "Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast". Nature. 340 (6230): 205–9. doi:10.1038/340205a0. PMID 2666856.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, Citovsky V (2001). "Genetic transformation of HeLa cells by Agrobacterium". Proc. Natl. Acad. Sci. U.S.A. 98 (4): 1871–6. doi:10.1073/pnas.041327598. PMC 29349. PMID 11172043.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Waters VL (2001). "Conjugation between bacterial and mammalian cells". Nat. Genet. 29 (4): 375–6. doi:10.1038/ng779. PMID 11726922.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Yoon YG, Koob MD (2005). "Transformation of isolated mammalian mitochondria by bacterial conjugation". Nucleic Acids Res. 33 (16): e139. doi:10.1093/nar/gni140. PMC 1201378. PMID 16157861.
External links
- Science project: Bacterial conjugation Transgenic poplars: Is horizontal gene transfer from Agrobacteria to endophytic bacteria possible?
- Bacterial conjugation (a Flash animation)
