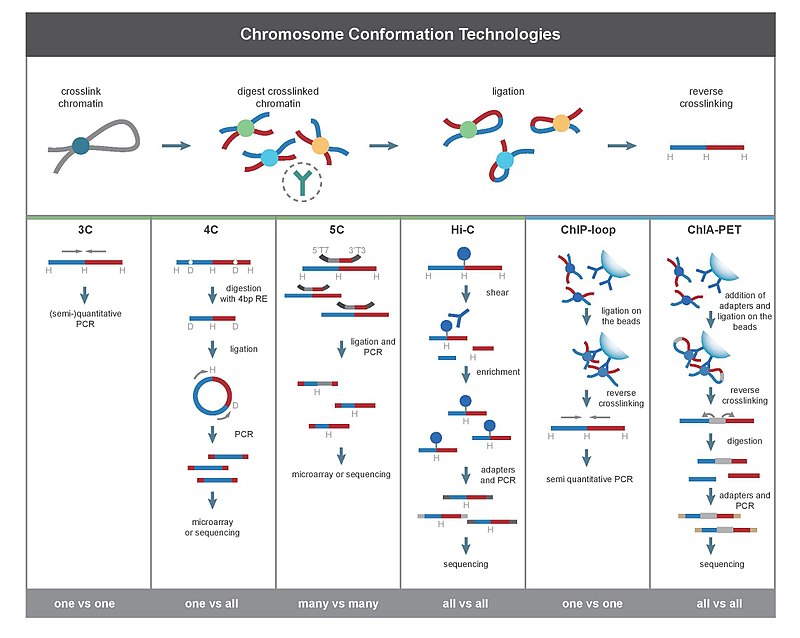
Chromosome conformation capture

Chromosome conformation capture techniques (often abbreviated to 3C technologies or 3C-based methods[1]) are a set of molecular biology methods used to analyze the spatial organization of chromatin in a cell. These methods quantify the number of interactions between genomic loci that are nearby in 3-D space, but may be separated by many nucleotides in the linear genome.[2] Such interactions may result from biological functions, such as promoter-enhancer interactions, or from random polymer looping, where undirected physical motion of chromatin causes loci to collide.[3] Interaction frequencies may be analyzed directly,[4] or they may be converted to distances and used to reconstruct 3-D structures.[5]
The chief difference between 3C-based methods is their scope. For example, when using PCR to detect interaction in a 3C experiment, the interactions between two specific fragments are quantified. In contrast, Hi-C quantifies interactions between all possible pairs of fragments simultaneously. Deep sequencing of material produced by 3C also produces genome-wide interactions maps.
History
Historically, microscopy was the primary method of investigating nuclear organization,[6] which can be dated back to 1590.[7]

In 1879, Walther Flemming coined the term chromatin.[8]
In 1883, August Weismann connected chromatin with heredity.
In 1884, Albrecht Kossel discovered histones.
In 1888, Sutton and Boveri proposed the theory of continuity of chromatin during the cell cycle [9]
In 1889, Wilhelm von Waldemeyer created the term "chromosome".[10]
In 1928, Emil Heitz coined the term Heterochromatin and Euchromatin.[11]
In 1942, Conrad Waddington postulated the epigenetic landscapes.[12]
In 1948, Rollin Hotchkiss discovered DNA methylation.[13]
In 1953, Watson and Crick discovered the double helix structure of DNA.[14]
In 1961, Mary Lyon postulated the principle of X-inactivation.
In 1973/1974, chromatin fiber was discovered.[12]
In 1975, Chambon coined the term nucleosomes.[12]
In 1982, Chromosome territories were discovered.[15]
In 1984, John T. Lis innovated the Chromatin immunoprecipitation technique.
In 1993, the Nuclear Ligation Assay was published, a method that could determine circularization frequencies of DNA in solution. This assay was used to show that estrogen induces an interaction between the prolactin gene promoter and a nearby enhancer.[16]
In 2002, Job Dekker introduced the new idea that dense matrices of interaction frequencies between loci could be used to infer the spatial organization of genomes. This idea was the basis for his development of the chromosome conformation capture (3C) assay, published in 2002 by Job Dekker and colleagues in the Kleckner lab at Harvard University.[17][18]
In 2003, the Human Genome Project was finished.
In 2006, Marieke Simonis invented 4C,[19] Dostie, in the Dekker lab, invented 5C.[20]
In 2007, B. Franklin Pugh innovated ChIP-seq technique.[21]
In 2009, Lieberman-Aiden, and Job Dekker invented Hi-C,[22] Melissa J. Fullwood invented ChIA-pet technique.[23]
In 2012, The Ren group, and the groups led by Edith Heard and Job Dekker discovered Topologically Associating Domains (TADs) in mammals.[24] Nora, E.P., Lajoie, B.R., Schulz, E.G., Giorgetti, L., Okamoto, I., Servant, N., Piolot, T., van Berkum, N.L., Meisig, J., Sedat, J., Gribnau, J., Barillot, E., Blüthgen, N., Dekker, J., Heard, E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 485(7398):381-385. https://doi.org/10.1038/nature11049</ref>
Experimental methods
All 3C methods start with a similar set of steps, performed on a sample of cells.
First, the cell genomes are cross-linked with formaldehyde,[25] which introduces bonds that "freeze" interactions between genomic loci. Treatment of cells with 1-3% formaldehyde, for 10-30min at room temperature is most common, however, standardization for preventing high protein-DNA cross linking is necessary, as this may negatively affect the efficiency of restriction digestion in the subsequent step.[26] The genome is then cut into fragments with a restriction endonuclease. The size of restriction fragments determines the resolution of interaction mapping. Restriction enzymes (REs) that make cuts on 6bp recognition sequences, such as EcoR1 or HindIII, are used for this purpose, as they cut the genome once every 4000bp, giving ~ 1 million fragments in the human genome.[26][27] For more precise interaction mapping, a 4bp recognizing RE may also be used. The next step is, random ligation. This takes place at low DNA concentrations in the presence of T4 DNA ligase,[28] such that ligation between cross-linked interacting fragments is favored over ligation between fragments that are not cross-linked. Subsequently, interacting loci are quantified by amplifying ligated junctions by PCR methods.[26][28]
Original methods
3C (one-vs-one)
The chromosome conformation capture (3C) experiment quantifies interactions between a single pair of genomic loci. For example, 3C can be used to test a candidate promoter-enhancer interaction. Ligated fragments are detected using PCR with known primers.[2][17]
4C (one-vs-all)
Chromosome conformation capture-on-chip (4C) captures interactions between one locus and all other genomic loci. It involves a second ligation step, to create self-circularized DNA fragments, which are used to perform inverse PCR. Inverse PCR allows the known sequence to be used to amplify the unknown sequence ligated to it.[2][29] In contrast to 3C and 5C, the 4C technique does not require the prior knowledge of both interacting chromosomal regions. Results obtained using 4C are highly reproducible with most of the interactions that are detected between regions proximal to one another. On a single microarray, approximately a million interactions can be analyzed. [citation needed]
5C (many-vs-many)
Chromosome conformation capture carbon copy (5C) detects interactions between all restriction fragments within a given region, with this region's size typically no greater than a megabase.[2][30] This is done by ligating universal primers to all fragments. However, 5C has relatively low coverage. The 5C technique overcomes the junctional problems at the intramolecular ligation step and is useful for constructing complex interactions of specific loci of interest. This approach is unsuitable for conducting genome-wide complex interactions since that will require millions of 5C primers to be used.[citation needed]
Hi-C (all-vs-all)
Hi-C uses high-throughput sequencing to find the nucleotide sequence of fragments.[2][31] The original protocol used paired end sequencing, which retrieves a short sequence from each end of each ligated fragment. As such, for a given ligated fragment, the two sequences obtained should represent two different restriction fragments that were ligated together in the random ligation step. The pair of sequences are individually aligned to the genome, thus determining the fragments involved in that ligation event. Hence, all possible pairwise interactions between fragments are tested.
Researchers attempt to study the extent of Hi-C's detection through a study focusing on screening primary brain tumours.[32] Prior to screening tumours, Hi-C was primarily focused on cell lines.[33]
Sequence capture-based methods
A number of methods use oligonucleotide capture to enrich 3C and Hi-C libraries for specific loci of interest.[34] These methods include Capture-C,[35] NG Capture-C,[36] Capture-3C,[37] and Capture Hi-C.[38] These methods are able to produce higher resolution and sensitivity than 4C based methods.[39]
Single-cell methods
Single-cell Hi-C can be used to investigate the interactions occurring in individual cells.[40][41]
Immunoprecipitation-based methods
ChIP-loop
ChIP-loop combines 3C with ChIP-seq to detect interactions between two loci of interest mediated by a protein of interest.[2][42] The ChIP-loop may be useful in identifying long-range cis-interactions and trans interaction mediated through proteins since frequent DNA collisions will not occur. [citation needed]
Genome wide methods
ChIA-PET combines Hi-C with ChIP-seq to detect all interactions mediated by a protein of interest.[2][43] HiChIP was designed to allow similar analysis as ChIA-PET with less input material.[44]
Biological impact
3C methods have led to a number of biological insights, including the discovery of new structural features of chromosomes, the cataloguing of chromatin loops, and increased understanding of transcriptional regulation mechanisms (the disruption of which can lead to disease).[6]
3C methods have demonstrated the importance of spatial proximity of regulatory elements to the genes that they regulate. For example, in tissues that express globin genes, the β-globin locus control region forms a loop with these genes. This loop is not found in tissues where the gene is not expressed.[45] This technology has further aided the genetic and epigenetic study of chromosomes both in model organisms and in humans.[not verified in body]
These methods have revealed large-scale organization of the genome into topologically associating domains (TADs), which correlate with epigenetic markers. Some TADs are transcriptionally active, while others are repressed.[46] Many TADs have been found in D. melanogaster, mouse and human.[47] Moreover, CTCF and cohesion play important roles in determining TADs and enhancer-promoter interactions. The result shows that the orientation of CTCF binding motifs in an enhancer-promoter loop should be facing to each other in order for the enhancer to find its correct target.[48]
Human disease
There are several diseases caused by defects in promoter-enhancer interactions, which is reviewed in this paper.[49]
Beta thalassemia is a certain type of blood disorders caused by a deletion of LCR enhancer element.[50][51]
Holoprosencephaly is cephalic disorder caused by a mutation in the SBE2 enhancer element, which in turn weakened the production of SHH gene.[52]
PPD2 (polydactyly of a triphalangeal thumb) is caused by a mutation of ZRS enhancer, which in turn strengthened the production of SHH gene.[53][54]
Adenocarcinoma of the lung can be caused by a duplication of enhancer element for MYC gene.[55]
T-cell acute lymphoblastic leukemia is caused by an introduction of a new enhancer.[56]
Data analysis
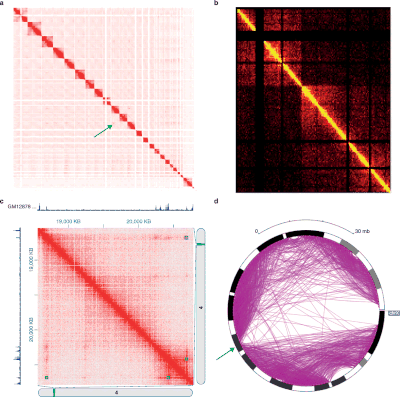
The different 3C-style experiments produce data with very different structures and statistical properties. As such, specific analysis packages exist for each experiment type.[62]
Hi-C data is often used to analyze genome-wide chromatin organization, such as topologically associating domains (TADs), linearly contiguous regions of the genome that are associated in 3-D space.[46] Several algorithms have been developed to identify TADs from Hi-C data.[4][63]
Hi-C and its subsequent analyses are evolving. Fit-Hi-C [3] is a method based on a discrete binning approach with modifications of adding distance of interaction (initial spline fitting, aka spline-1) and refining the null model (spline-2). The result of Fit-Hi-C is a list of pairwise intra-chromosomal interactions with their p-values and q-values. Hi-C data visualization tools are recently reviewed in Gurken et al.
The 3-D organization of the genome can also be analyzed via eigendecomposition of the contact matrix. Each eigenvector corresponds to a set of loci, which are not necessarily linearly contiguous, that share structural features.[64]
A significant confounding factor in 3C technologies is the frequent non-specific interactions between genomic loci that occur due to random polymer behavior. An interaction between two loci must be confirmed as specific through statistical significance testing.[3]
DNA motif analysis
DNA motifs are specific short DNA sequences, often 8-20 nucleotides in length,[65] which are statistically overrepresented in a set of sequences with a common biological function. Currently, regulatory motifs on the long-range chromatin interactions have not been studied extensively. Several studies have focused on elucidating the impact of DNA motifs in promoter-enhancer interactions.
Guo et al. has identified the orientation of two CTCF motifs in the promoter-enhancer border sequences is very crucial for targeting the right gene; the two CTCF motifs have to face to each other.[66]
Bailey et al. has identified that ZNF143 motif in the promoter regions provides sequence specificity for promoter-enhancer interactions.[67] Mutation of ZNF143 motif decreased the frequency of promoter-enhancer interactions suggesting that ZNF143 is a novel chromatin-looping factor.
For genome-scale motif analysis, in 2016, Wong et al. reported a list of 19,491 DNA motif pairs for K562 cell line on the promoter-enhancer interactions.[68] As a result, they claimed that motif pairing multiplicity (number of motifs that are paired with a given motif) is linked to interaction distance and regulatory region type. In the next year, Wong published another article reporting 18,879 motif pairs in 6 human cell lines.[69] A novel contribution of this work is MotifHyades (written in Matlab), a motif discovery tool that can be directly applied to paired sequences.
Cancer genome analysis
The 3C-based techniques can provide insights into the chromosomal rearrangements in the cancer genomes.[70] Moreover, they can show changes of spatial proximity for regulatory elements and their target genes, which bring deeper understanding of the structural and functional basis of the genome.[71]
Taberlay et al. studied the disruption of 3D genome organization in the context of prostate cancer.[72] Copy-number variation, long-range epigenetic remodeling, and atypical gene expression programs were analyzed. Specifically, they found a bifurcation of a single TAD (normal) into 2 distinct smaller TADs (cancer) caused by a common deletion on 17p13.1. The data can be access in the GEO database with GSE73785.
Harewood el. al proposed to use Hi-C as a tool to detect chromosomal rearrangements and copy number variations.[70] The cancer dataset consists of 6 types of brain tumors, 2 lymphoblastic cell lines, and 1 control cell line. The GEO accession number is GSE81879.
Ferhat Ay et al. analyzed 10 different cancer cell lines to devise a new set of algorithms, including identification of copy number variations (HiCnv), inter-chromosomal translocations (HiCtrans), and simulation of Hi-C data (AveSim).[73] The datasets are from ENCODE project.[74]
Luo et al. performed Hi-C experiment and found a prostate cancer risk region (7p15.2) in an anchor point for a repressive chromatin interaction that contains HOXA13 gene.[75] The deletion of the 3p15.2 locus can upregulate genes in the HOXA locus. The GEO accession number is GSE98898.
Dixon et al. designed an algorithm that can use Hi-C data to detect SV events in 35 cancer cell lines, 16 of which are from the ENCODE project,[74] including A549, ACHN, Caki2, DLD1, G401, HeLa-S3, HepG2, LNCaP clone FGC, NCI-H460, PANC-1, RPMI-7951, SJCRH30, SK-MEL-5, SK-N-DZ, SK-N-MC, T-47D.[76]
References
- ^ de Wit, E.; de Laat, W. (3 January 2012). "A decade of 3C technologies: insights into nuclear organization". Genes & Development. 26 (1): 11–24. doi:10.1101/gad.179804.111. PMC 3258961. PMID 22215806.
- ^ a b c d e f g Hakim, Ofir (2012). "SnapShot: Chromosome confirmation capture". Cell. 148 (5): 1068.e1–2. doi:10.1016/j.cell.2012.02.019. PMID 22385969.
- ^ a b c Ay, F.; Bailey, T. L.; Noble, W. S. (5 February 2014). "Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts". Genome Research. 24 (6): 999–1011. doi:10.1101/gr.160374.113. PMC 4032863. PMID 24501021.
- ^ a b Rao, Suhas S.P.; Huntley, Miriam H.; Durand, Neva C.; Stamenova, Elena K.; Bochkov, Ivan D.; Robinson, James T.; Sanborn, Adrian L.; Machol, Ido; Omer, Arina D.; Lander, Eric S.; Aiden, Erez Lieberman (December 2014). "A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping". Cell. 159 (7): 1665–1680. doi:10.1016/j.cell.2014.11.021. PMC 5635824. PMID 25497547.
- ^ Varoquaux, N.; Ay, F.; Noble, W. S.; Vert, J.-P. (15 June 2014). "A statistical approach for inferring the 3D structure of the genome". Bioinformatics. 30 (12): i26–i33. doi:10.1093/bioinformatics/btu268. PMC 4229903. PMID 24931992.
- ^ a b Denker, Annette; de Laat, Wouter (23 June 2016). "The second decade of 3C technologies: detailed insights into nuclear organization". Genes & Development. 30 (12): 1357–1382. doi:10.1101/gad.281964.116. PMC 4926860. PMID 27340173.
- ^ http://www.history-of-the-microscope.org/history-of-the-microscope-who-invented-the-microscope.php
- ^ "Photography by Benjamin Saur Tübingen Walther Flemming a German physician and".
- ^ MARTINS, L.A.-C.P.. Did Sutton and Boveri propose the so-called Sutton-Boveri chromosome hypothesis?. Genet. Mol. Biol. [online]. 1999, vol.22, n.2 [cited 2017-12-07], pp.261-272. Available from: <http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1415-47571999000200022&lng=en&nrm=iso>. ISSN 1415-4757. https://dx.doi.org/10.1590/S1415-47571999000200022.
- ^ "Genes and genetics: The language of scientific discovery". 2012-08-16.
- ^ "Heterochromatin and euchromatin mains". 2015-02-05.
- ^ a b c Deichmann, U. (2016). Epigenetics: The origins and evolution of a fashionable topic. Developmental Biology, 416(1), 249–254. https://doi.org/https://doi.org/10.1016/j.ydbio.2016.06.005
- ^ Lu, H., Liu, X., Deng, Y., & Qing, H. (2013). DNA methylation, a hand behind neurodegenerative diseases. Frontiers in Aging Neuroscience, 5, 85. http://doi.org/10.3389/fnagi.2013.00085
- ^ "The Francis Crick Papers: The Discovery of the Double Helix, 1951-1953".
- ^ Cremer, T., & Cremer, M. (2010). Chromosome Territories. Cold Spring Harbor Perspectives in Biology, 2(3), a003889. http://doi.org/10.1101/cshperspect.a003889
- ^ Cullen, K.; Kladde, M.; Seyfred, M. (9 July 1993). "Interaction between transcription regulatory regions of prolactin chromatin". Science. 261 (5118): 203–206. doi:10.1126/science.8327891.
- ^ a b Dekker, J; Rippe, K; Dekker, M; Kleckner, N (15 February 2002). "Capturing chromosome conformation". Science. 295 (5558): 1306–11. doi:10.1126/science.1067799. PMID 11847345.
- ^ Osborne, CS; Ewels, PA; Young, AN (January 2011). "Meet the neighbours: tools to dissect nuclear structure and function". Briefings in Functional Genomics. 10 (1): 11–7. doi:10.1093/bfgp/elq034. PMC 3080762. PMID 21258046.
- ^ Simonis, M., Klous, P., Splinter, E., Moshkin, Y., Willemsen, R., & de Wit, E. (2006). Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet, 38. https://doi.org/10.1038/ng1896
- ^ Dostie, J., Richmond, T. A., Arnaout, R. A., Selzer, R. R., Lee, W. L., & Honan, T. A. (2006). Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res, 16. https://doi.org/10.1101/gr.5571506
- ^ Albert, I., Mavrich, T. N., Tomsho, L. P., Qi, J., Zanton, S. J., Schuster, S. C., & Pugh, B. F. (2007). Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature, 446, 572. Retrieved from https://dx.doi.org/10.1038/nature05632
- ^ Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., & Telling, A. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 326. https://doi.org/10.1126/science.1181369
- ^ Fullwood, M. J., Liu, M. H., Pan, Y. F., Liu, J., Xu, H., & Mohamed, Y. B. (2009). An oestrogen-receptor-alpha-bound human chromatin interactome. Nature, 462. https://doi.org/10.1038/nature08497
- ^ Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., … Ren, B. (2012). Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature, 485(7398), 376–380. http://doi.org/10.1038/nature11082
- ^ Gavrilov, Alexey; Eivazova, Elvira; Pirozhkova, Iryna; Lipinski, Marc; Razin, Sergey; Vassetzky, Yegor (2009). Chromatin Immunoprecipitation Assays. Methods in Molecular Biology. Vol. 567. Humana Press, Totowa, NJ. pp. 171–188. doi:10.1007/978-1-60327-414-2_12. ISBN 9781603274135. PMID 19588093.
- ^ a b c Naumova, Natalia; Smith, Emily M.; Zhan, Ye; Dekker, Job (2012). "Analysis of long-range chromatin interactions using Chromosome Conformation Capture". Methods. 58 (3): 192–203. doi:10.1016/j.ymeth.2012.07.022. PMC 3874837. PMID 22903059.
- ^ Belton, Jon-Matthew; Dekker, Job (2015-06-01). "Chromosome Conformation Capture (3C) in Budding Yeast". Cold Spring Harbor Protocols. 2015 (6): 580–6. doi:10.1101/pdb.prot085175. ISSN 1940-3402. PMID 26034304.
- ^ a b Gavrilov, Alexey A.; Golov, Arkadiy K.; Razin, Sergey V. (2013-03-26). "Actual Ligation Frequencies in the Chromosome Conformation Capture Procedure". PLOS ONE. 8 (3): e60403. doi:10.1371/journal.pone.0060403. ISSN 1932-6203. PMC 3608588. PMID 23555968.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Simonis, Marieke (2006). "Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture–on-chip (4C)". Nature Genetics. 38 (11): 1348–1354. doi:10.1038/ng1896. PMID 17033623. Retrieved 30 May 2016.
- ^ Dostie, Josee (2006). "Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements". Genome Research. 16 (10): 1299–1309. doi:10.1101/gr.5571506. PMC 1581439. PMID 16954542.
- ^ Lieberman-Aiden, Erez (2009). "Comprehensive mapping of long-range interactions reveals folding principles of the human genome". Science. 326 (5950): 289–293. doi:10.1126/science.1181369. PMC 2858594. PMID 19815776. Retrieved 30 May 2016.
- ^ Harewood, Louise; Kishore, Kamal; Eldridge, Matthew D.; Wingett, Steven; Pearson, Danita; Schoenfelder, Stefan; Collins, V. Peter; Fraser, Peter (2017-06-27). "Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours". Genome Biology. 18 (1): 125. doi:10.1186/s13059-017-1253-8. ISSN 1474-760X. PMC 5488307. PMID 28655341.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Burton, Joshua N.; Adey, Andrew; Patwardhan, Rupali P.; Qiu, Ruolan; Kitzman, Jacob O.; Shendure, Jay (December 2013). "Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions". Nature Biotechnology. 31 (12): 1119–1125. doi:10.1038/nbt.2727. ISSN 1546-1696. PMC 4117202. PMID 24185095.
- ^ Schmitt, Anthony D.; Hu, Ming; Ren, Bing (1 September 2016). "Genome-wide mapping and analysis of chromosome architecture". Nature Reviews Molecular Cell Biology. 17 (12): 743–755. doi:10.1038/nrm.2016.104. PMC 5763923. PMID 27580841.
- ^ Hughes, Jim R; Roberts, Nigel; McGowan, Simon; Hay, Deborah; Giannoulatou, Eleni; Lynch, Magnus; De Gobbi, Marco; Taylor, Stephen; Gibbons, Richard (2014-01-12). "Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment". Nature Genetics. 46 (2): 205–212. doi:10.1038/ng.2871. ISSN 1061-4036. PMID 24413732.
- ^ Davies, James O J; Telenius, Jelena M; McGowan, Simon J; Roberts, Nigel A; Taylor, Stephen; Higgs, Douglas R; Hughes, Jim R (2015-11-23). "Multiplexed analysis of chromosome conformation at vastly improved sensitivity". Nature Methods. 13 (1): 74–80. doi:10.1038/nmeth.3664. ISSN 1548-7091. PMC 4724891. PMID 26595209.
- ^ Hughes, Jim (2014). "Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment". Nature Genetics. 46 (2): 205–212. doi:10.1038/ng.2871. PMID 24413732. Retrieved 6 June 2016.
- ^ Jäger, Roland; Migliorini, Gabriele; Henrion, Marc; Kandaswamy, Radhika; Speedy, Helen E.; Heindl, Andreas; Whiffin, Nicola; Carnicer, Maria J.; Broome, Laura; Dryden, Nicola; Nagano, Takashi; Schoenfelder, Stefan; Enge, Martin; Yuan, Yinyin; Taipale, Jussi; Fraser, Peter; Fletcher, Olivia; Houlston, Richard S. (19 February 2015). "Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci". Nature Communications. 6: 6178. doi:10.1038/ncomms7178. PMC 4346635. PMID 25695508.
- ^ Davies, James O J; Oudelaar, A Marieke; Higgs, Douglas R; Hughes, Jim R (2017-02). "How best to identify chromosomal interactions: a comparison of approaches". Nature Methods. 14 (2): 125–134. doi:10.1038/nmeth.4146. ISSN 1548-7091. PMID 28139673.
{{cite journal}}: Check date values in:|date=(help) - ^ Nagano, Takashi; Lubling, Yaniv; Stevens, Tim J.; Schoenfelder, Stefan; Yaffe, Eitan; Dean, Wendy; Laue, Ernest D.; Tanay, Amos; Fraser, Peter (25 September 2013). "Single-cell Hi-C reveals cell-to-cell variability in chromosome structure". Nature. 502 (7469): 59–64. doi:10.1038/nature12593. PMC 3869051. PMID 24067610.
- ^ Schwartzman, Omer; Tanay, Amos (13 October 2015). "Single-cell epigenomics: techniques and emerging applications". Nature Reviews Genetics. 16 (12): 716–726. doi:10.1038/nrg3980. PMID 26460349.
- ^ Horike, Shin-ichi (2005). "Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome". Nature Genetics. 37 (1): 31–40. doi:10.1038/ng1491. PMID 15608638. Retrieved 30 May 2016.
- ^ Fullwood, Melissa (2009). "An oestrogen-receptor-alpha-bound human chromatin interactome". Nature. 462 (7269): 58–64. doi:10.1038/nature08497. PMC 2774924. PMID 19890323. Retrieved 30 May 2016.
- ^ Mumbach, Maxwell R; Rubin, Adam J; Flynn, Ryan A; Dai, Chao; Khavari, Paul A; Greenleaf, William J; Chang, Howard Y (2016-09-19). "HiChIP: efficient and sensitive analysis of protein-directed genome architecture". Nature Methods. 13 (11): 919–922. doi:10.1038/nmeth.3999. ISSN 1548-7105. PMC 5501173. PMID 27643841.
- ^ Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002). "Looping and interaction between hypersensitive sites in the active beta-globin locus". Mol. Cell. 10 (6): 1453–1465. doi:10.1016/S1097-2765(02)00781-5. PMID 12504019.
- ^ a b Cavalli, Giacamo (2013). "Functional implications of genome topology". Nature Structural & Molecular Biology. 20 (3): 290–299. doi:10.1038/nsmb.2474. PMID 23463314. Retrieved 12 June 2016.
- ^ J. Dekker, M. A. Marti-Renom, and L. A. Mirny, “Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data,” Nature reviews. Genetics, vol. 14, no. 6. pp. 390–403, Jun-2013.
- ^ Y. Guo et al., “CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function.,” Cell, vol. 162, no. 4, pp. 900–910, Aug. 2015
- ^ Krijger, P. H. L., & de Laat, W. (2016). Regulation of disease-associated gene expression in the 3D genome. Nat Rev Mol Cell Biol, 17(12), 771–782. Retrieved from https://dx.doi.org/10.1038/nrm.2016.138
- ^ Fritsch, E. F., Lawn, R. M. & Maniatis, T. Characterisation of deletions which affect the expression of fetal globin genes in man. Nature 279, 598–603 (1979)
- ^ Van der Ploeg, L. H. et al. γ-Β-Thalassaemia studies showing that deletion of the γ- and δ-genes influences β-globin gene expression in man. Nature 283, 637–642 (1980).
- ^ Jeong, Y., El-Jaick, K., Roessler, E., Muenke, M. & Epstein, D. J. A functional screen for sonic hedgehog regulatory elements across a 1Mb interval identifies long-range ventral forebrain enhancers. Development 133, 761–772 (2006)
- ^ Lettice, L. A. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 (2003)
- ^ Wieczorek, D. et al. A specific mutation in the distant sonic hedgehog (SHH) cis-regulator (ZRS) causes Werner mesomelic syndrome (WMS) while complete ZRS duplications underlie Haas type polysyndactyly and preaxial polydactyly (PPD) with or without triphalangeal thumb. Hum. Mutat. 31, 81–89 (2010).
- ^ Zhang, X. et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 48, 176–182 (2016)
- ^ Mansour, M. R. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377 (2014).
- ^ Lajoie, Bryan R; van Berkum, Nynke L; Sanyal, Amartya; Dekker, Job (1 October 2009). "My5C: web tools for chromosome conformation capture studies". Nature Methods. 6 (10): 690–691. doi:10.1038/nmeth1009-690. PMC 2859197. PMID 19789528.
- ^ Deng, Xinxian; Ma, Wenxiu; Ramani, Vijay; Hill, Andrew; Yang, Fan; Ay, Ferhat; Berletch, Joel B.; Blau, Carl Anthony; Shendure, Jay; Duan, Zhijun; Noble, William S.; Disteche, Christine M. (7 August 2015). "Bipartite structure of the inactive mouse X chromosome". Genome Biology. 16 (1): 152. doi:10.1186/s13059-015-0728-8. PMC 4539712. PMID 26248554.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Rao, Suhas S.P.; Huntley, Miriam H.; Durand, Neva C.; Stamenova, Elena K.; Bochkov, Ivan D.; Robinson, James T.; Sanborn, Adrian L.; Machol, Ido; Omer, Arina D.; Lander, Eric S.; Aiden, Erez Lieberman (December 2014). "A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping". Cell. 159 (7): 1665–1680. doi:10.1016/j.cell.2014.11.021. PMC 5635824. PMID 25497547.
- ^ Zhou, Xin; Lowdon, Rebecca F; Li, Daofeng; Lawson, Heather A; Madden, Pamela A F; Costello, Joseph F; Wang, Ting (29 April 2013). "Exploring long-range genome interactions using the WashU Epigenome Browser". Nature Methods. 10 (5): 375–376. doi:10.1038/nmeth.2440. PMC 3820286. PMID 23629413.
- ^ Yardımcı, Galip Gürkan; Noble, William Stafford (3 February 2017). "Software tools for visualizing Hi-C data". Genome Biology. 18 (1): 26. doi:10.1186/s13059-017-1161-y. PMC 5290626. PMID 28159004.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Schmitt, AD; Hu, M; Ren, B (December 2016). "Genome-wide mapping and analysis of chromosome architecture". Nature Reviews Molecular Cell Biology. 17 (12): 743–755. doi:10.1038/nrm.2016.104. PMC 5763923. PMID 27580841.
- ^ Dixon, Jesse R.; Selvaraj, Siddarth; Yue, Feng; Kim, Audrey; Li, Yan; Shen, Yin; Hu, Ming; Liu, Jun S.; Ren, Bing (11 April 2012). "Topological domains in mammalian genomes identified by analysis of chromatin interactions". Nature. 485 (7398): 376–380. doi:10.1038/nature11082. PMC 3356448. PMID 22495300.
- ^ Imakaev, Maxim; Fudenberg, Geoffrey; McCord, Rachel Patton; Naumova, Natalia; Goloborodko, Anton; Lajoie, Bryan R; Dekker, Job; Mirny, Leonid A (2 September 2012). "Iterative correction of Hi-C data reveals hallmarks of chromosome organization". Nature Methods. 9 (10): 999–1003. doi:10.1038/nmeth.2148. PMC 3816492. PMID 22941365.
- ^ F. Zambelli, G. Pesole, and G. Pavesi, “Motif discovery and transcription factor binding sites before and after the next-generation sequencing era.,” Brief. Bioinform., vol. 14, no. 2, pp. 225–37, Mar. 2013
- ^ Guo Y, Xu Q, Canzio D, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162(4):900-910. doi:10.1016/j.cell.2015.07.038.
- ^ Bailey, S. D., Zhang, X., Desai, K., Aid, M., Corradin, O., Cowper-Sal·lari, R., … Lupien, M. (2015). ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nature Communications, 2, 6186. Retrieved from https://dx.doi.org/10.1038/ncomms7186
- ^ K. Wong, Y. Li, and C. Peng, “Identification of coupling DNA motif pairs on long-range chromatin interactions in human,” vol. 32, no. September 2015, pp. 321–324, 2016.
- ^ Ka-Chun Wong; MotifHyades: expectation maximization for de novo DNA motif pair discovery on paired sequences, Bioinformatics, Volume 33, Issue 19, 1 October 2017, Pages 3028–3035, https://doi.org/10.1093/bioinformatics/btx381
- ^ a b L. Harewood et al., “Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours,” pp. 1–11, 2017.
- ^ P. C. Taberlay et al., “Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations.,” Genome Res., vol. 26, no. 6, pp. 719–731, Jun. 2016.
- ^ Taberlay, P. C., Achinger-Kawecka, J., Lun, A. T. L., Buske, F. A., Sabir, K., Gould, C. M., … Clark, S. J. (2016). Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Research, 26(6), 719–731. https://doi.org/10.1101/gr.201517.115
- ^ A. Chakraborty and F. Ay, “Identification of copy number variations and translocations in cancer cells from Hi-C data,” 2017.
- ^ a b "ENCODE: Encyclopedia of DNA Elements – ENCODE".
- ^ Luo, Z., Rhie, S. K., Lay, F. D., & Farnham, P. J. (2017). A Prostate Cancer Risk Element Functions as a Repressive Loop that Regulates HOXA13. Cell Reports, 21(6), 1411–1417. https://doi.org/10.1016/j.celrep.2017.10.048
- ^ Dixon, J., et al. (2018). https://www.nature.com/articles/s41588-018-0195-8
Further reading
- Barutcu AR, Fritz AJ, Zaidi SK, van Wijnen AJ, Lian JB, Stein JL, Nickerson JA, Imbalzano AN, Stein GS (January 2016). "C-ing the Genome: A Compendium of Chromosome Conformation Capture Methods to Study Higher-Order Chromatin Organization". J. Cell. Physiol. 231 (1): 31–35. doi:10.1002/jcp.25062. PMC 4586368. PMID 26059817.
- Marbouty M, Koszul R (December 2015). "Metagenome Analysis Exploiting High-Throughput Chromosome Conformation Capture (3C) Data" (review). Trends Genet. 31 (12): 673–682. doi:10.1016/j.tig.2015.10.003. PMID 26608779. Retrieved 23 April 2016.
{{cite journal}}: Unknown parameter|subscription=ignored (|url-access=suggested) (help) - Dekker J. (25 November 2014). "Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture". Epigenetics & Chromatin. 7 (1): 25. doi:10.1186/1756-8935-7-25. PMC 4247682. PMID 25435919.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - O'Sullivan JM, Hendy MD, Pichugina T, Wake GC, Langowski J (September–October 2013). "The statistical-mechanics of chromosome conformation capture". Nucleus. 4 (5): 390–398. doi:10.4161/nucl.26513. PMC 3899129. PMID 24051548.
- Umbarger MA (November 2012). "Chromosome conformation capture assays in bacteria" (review). Methods. 58 (3): 212–20. doi:10.1016/j.ymeth.2012.06.017. PMID 22776362. Retrieved 23 April 2016.
{{cite journal}}: Unknown parameter|subscription=ignored (|url-access=suggested) (help) - Gavrilov A, Eivazova E, Priozhkova I, Lipinski M, Razin S, Vassetzky Y (2009). Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods in Molecular Biology. Vol. 567. pp. 171–188. doi:10.1007/978-1-60327-414-2_12. ISBN 978-1-60327-413-5. PMID 19588093.
{{cite book}}:|format=requires|url=(help);|journal=ignored (help) See also * Gavrilov A, Eivazova E, Priozhkova I, Lipinski M, Razin S, Vassetzky Y (2009). ibid., erratum. Methods in Molecular Biology. Vol. 567. pp. 171–188. doi:10.1007/978-1-60327-414-2_12. ISBN 978-1-60327-413-5. PMID 19588093.{{cite book}}:|journal=ignored (help)[full citation needed] - Parelho, Vania; Merkenschlager, Matthias (2005). "Gene Expression: Growing up together may help genes go their separate ways" (news and commentary). European Journal of Human Genetics. 13 (July 6): 993–994. doi:10.1038/sj.ejhg.5201464. PMID 15999115. Retrieved 23 April 2016.
- Marvin, Marcus; Tan-Wong, Sue Mei (2016-04-23). "Chromosome conformation capture" (commercial method). abcam.com. Retrieved 23 April 2016.