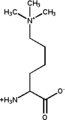
Methyllysine
Methyllysine is derivative of the amino acid residue lysine where the sidechain ammonium group has been methylated one or more times.
Such methylated lysines play an important role in epigenetics; the methylation of specific lysines of certain histones in a nucleosome alters the binding of the surrounding DNA to those histones, which in turn affects the expression of genes on that DNA.[1][2] The binding is affected because the effective radius of the positive charge is increased (methyl groups are larger than the hydrogen atoms they replace), reducing the strongest potential electrostatic attraction with the negatively charged DNA. Moreover, the methyl groups are themselves hydrophobic, and alter the structure of water in their vicinity, similar to tetramethyl ammonium.[3]
It is thought that the methylation of lysine (and arginine) on histone tails does not directly affect their binding to DNA. Rather, such methyl marks recruit other proteins that modulate chromatin structure.[4]
In Protein Data Bank files, methylated lysines are indicated by the MLY or MLZ acronyms.
-
Unmethylated lysine
-
Monomethylated: 6-N-methyllysine
-
Dimethylated: (6-N,6-N)dimethyllysine
-
Trimethylated: (6-N,6-N,6-N)trimethyllysine
References
- ^ Ruthenburg AJ, Allis CD, Wysocka J (January 2007). "Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark". Molecular Cell. 25 (1): 15–30. doi:10.1016/j.molcel.2006.12.014. PMID 17218268.
- ^ Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (August 2003). "Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains". Genes & Development. 17 (15): 1870–81. doi:10.1101/gad.1110503. PMC 196235. PMID 12897054.
- ^ Finney JL, Soper AK, Turner JZ (1993). "Water perturbation close to non-polar groups in aqueous solutions" (PDF). Pure and Applied Chemistry. 65 (12): 2521–6.
TMA [Tetramethylammonium] hydrates as an apolar molecule: there is evidence for a cage-like average hydration structure, but one which is defective and disordered
- ^ Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M (April 2012). "Epigenetic protein families: a new frontier for drug discovery". Nature Reviews. Drug Discovery. 11 (5): 384–400. doi:10.1038/nrd3674. PMID 22498752.