SAM-II riboswitch
| SAM riboswitch (alpha-proteobacteria) | |
|---|---|
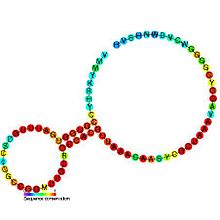 Predicted secondary structure and sequence conservation of SAM_alpha | |
| Identifiers | |
| Symbol | SAM_alpha |
| Rfam | RF00521 |
| Other data | |
| RNA type | Cis-reg; riboswitch |
| Domain(s) | Bacteria |
| GO | GO:0045814 |
| SO | SO:0000035 |
| PDB structures | PDBe |
The SAM-II riboswitch is a RNA element found predominantly in alpha-proteobacteria that binds S-adenosyl methionine (SAM).[1] Its structure and sequence appear to be unrelated to the SAM riboswitch found in Gram-positive bacteria. This SAM riboswitch is located upstream of the metA and metC genes in Agrobacterium tumefaciens, and other methionine and SAM biosynthesis genes in other alpha-proteobacteria. Like the other SAM riboswitch, it probably functions to turn off expression of these genes in response to elevated SAM levels. A significant variant of SAM-II riboswitches was found in Pelagibacter ubique and related marine bacteria and called SAM-V.[2] Also, like many structured RNAs, SAM-II riboswitches can tolerate long loops between their stems.[3]
Structure
The SAM-II riboswitch is short with less than 70 nucleotides and is structurally relatively simple being composed of a single hairpin and a pseudoknot.
See also
References
- ^ Corbino KA, Barrick JE, Lim J, et al. (2005). "Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria". Genome Biol. 6 (8): R70. doi:10.1186/gb-2005-6-8-r70. PMC 1273637. PMID 16086852.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Poiata E, Meyer MM, Ames TD, Breaker RR (November 2009). "A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria". RNA. 15 (11): 2046–56. doi:10.1261/rna.1824209. PMC 2764483. PMID 19776155.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Weinberg Z, Wang JX, Bogue J, et al. (March 2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol. 11 (3): R31. doi:10.1186/gb-2010-11-3-r31. PMC 2864571. PMID 20230605.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
External links
