Tissue typing
Tissue typing is a procedure in which the tissues of a prospective donor and recipient are tested for compatibility prior to transplantation. Mismatched donor and recipient tissues can lead to rejection of the tissues. There are multiple methods of tissue typing.
Overview
[edit]
During tissue typing, an individual's human leukocyte antigens (HLA) are identified.[1] HLA molecules are presented on the surface of cells and facilitate interactions between immune cells (such as dendritic cells and T cells) that lead to adaptive immune responses.[2] If HLA from the donor is recognized by the recipient's immune system as different from the recipient's own HLA, an immune response against the donor tissues can be triggered.[3] More specifically, HLA mismatches between organ donors and recipients can lead to the development of anti-HLA donor-specific antibodies (DSAs).[4] DSAs are strongly associated with the rejection of donor tissues in the recipient, and their presence is considered an indicator of antibody-mediated rejection.[5] When donor and recipient HLA are matched, donor tissues are significantly more likely to be accepted by the recipient's immune system.[3] During tissue typing, a number of HLA genes should be typed in both the donor and recipient, including HLA Class I A, B, and C genes, as well as HLA Class II DRB1, DRB3, DRB4, DRB5, DQA1, DQB1, DPA1, and DPB1 genes.[6] HLA typing is made more difficult by the fact that the HLA region is the most genetically variable region in the human genome.[7]
Methods of tissue typing
[edit]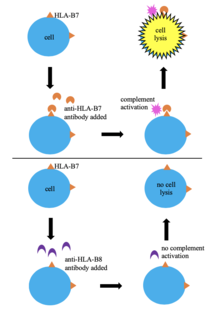
One of the first methods of tissue typing was through serological typing. In this technique, a donor's blood cells are HLA typed by mixing them with serum containing anti-HLA antibodies. If the antibodies recognize their epitope on the donor's HLA then complement activation occurs leads to cell lysis and death, allowing the cells to take up a dye (trypan blue). This allows for identification of the cells' HLA based indirectly on the specificity of the known antibodies in the serum. This method has been used widely since it is simple, quick, and low-cost; however, the huge variability in HLA alleles means that serum containing antibodies specific to the HLA of the cells being tested may not be available.[6][3] Serological typing does not give a clear picture of the HLA region and does not always result in successful HLA typing, so many laboratories have stopped using it in favor of more effective methods.[6][8]
Recently, other more effective approaches have emerged, including the use of polymerase chain reaction (PCR) based on sequence-specific primers (SSP) or sequence-specific oligonucleotide probes (SSOP).[3][6] However, SSP-PCR can be both time and resource consuming.[8] SSOP-PCR is better for HLA typing large numbers of individuals, for example, large numbers of donors for bone marrow registries.[8] RT-PCR is another approach to HLA typing that is fast and versatile, but it is expensive.[6]
Reference strand-mediated conformational analysis (RSCA) is yet another method used for HLA typing. In this method, an unknown HLA sample is mixed with a reference allele and run in a gel by electrophoresis.[8] RSCA is limited by the number of HLA reference alleles available since the HLA region is so diverse.[8]
Direct DNA sequencing is currently considered the best method of HLA typing, either by Sanger sequencing or next generation sequencing, though it can also be time-consuming and is one of the more expensive methods.[6][8] RNA sequencing can also be used, but many labs do not as RNA is unstable and prone to degradation.[8]
See also
[edit]References
[edit]- ^ Mulley, William R.; Hudson, Fiona; Lee, Darren; Holdsworth, Rhonda F. (2019). "Tissue typing for kidney transplantation for the general nephrologist". Nephrology. 24 (10): 997–1000. doi:10.1111/nep.13637. ISSN 1440-1797. PMID 31335997.
- ^ Nazahah, M.; Koh, M. B. C. (2015). "Tissue typing and its role in transplantation". ISBT Science Series. 10 (S1): 115–123. doi:10.1111/voxs.12168. ISSN 1751-2824. S2CID 71558083.
- ^ a b c d Mahdi, Batool Mutar (2013-02-23). "A glow of HLA typing in organ transplantation". Clinical and Translational Medicine. 2 (1): 6. doi:10.1186/2001-1326-2-6. ISSN 2001-1326. PMC 3598844. PMID 23432791.
- ^ Argani, Hassan (January 2019). "Anti-HLA Antibody: The Role of Epitopes in Organ Transplantation". Experimental and Clinical Transplantation. 17 (Suppl 1): 38–42. doi:10.6002/ect.MESOT2018.L41. PMID 30777521. S2CID 73478179.
- ^ Zhang, Rubin (2018-01-06). "Donor-Specific Antibodies in Kidney Transplant Recipients". Clinical Journal of the American Society of Nephrology. 13 (1): 182–192. doi:10.2215/CJN.00700117. ISSN 1555-9041. PMC 5753302. PMID 28446536.
- ^ a b c d e f Deaglio, Silvia; Amoroso, Antonio; Rinaldi, Mauro; Boffini, Massimo (2020-02-13). "HLA typing in lung transplantation: does high resolution fit all?". Annals of Translational Medicine. 8 (3): 45. doi:10.21037/atm.2020.01.45. ISSN 2305-5847. PMC 7036624. PMID 32154803.
- ^ Kishore, Amit; Petrek, Martin (2018). "Next-Generation Sequencing Based HLA Typing: Deciphering Immunogenetic Aspects of Sarcoidosis". Frontiers in Genetics. 9: 503. doi:10.3389/fgene.2018.00503. ISSN 1664-8021. PMC 6210504. PMID 30410504.
- ^ a b c d e f g Dunn, P. P. J. (2011). "Human leucocyte antigen typing: techniques and technology, a critical appraisal". International Journal of Immunogenetics. 38 (6): 463–473. doi:10.1111/j.1744-313X.2011.01040.x. ISSN 1744-313X. PMID 22059555. S2CID 44343932.
External links
[edit]- How Preimplantation tissue typing works, HFEA website
- Tissue+typing at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Understanding Tissue Typing
