V(D)J recombination
V(D)J recombination is a mechanism of genetic recombination that occurs in vertebrates, which randomly selects and assembles segments of genes encoding specific proteins with important roles in the immune system.[1] This site-specific recombination reaction generates a diverse repertoire of T cell receptor (TCR) and immunoglobulin (Ig) molecules that are necessary for the recognition of diverse antigens from bacterial, viral, and parasitic invaders, and from dysfunctional cells such as tumor cells.[2]
Background
Human antibody molecules (and B cell receptors) comprise heavy and light chains with both constant (C) and variable (V) regions that are encoded by three types of genes.
- Heavy chain gene – located on chromosome 14
- Light chain kappa (κ) gene – located on chromosome 2
- Light chain lambda (λ) gene – located chromosome 22
Multiple genes for the variable regions are encoded in the human genome that contain three distinct types of segments. For example, the immunoglobulin heavy chain region contains 65 Variable (V) genes[citation needed] plus 27 Diversity (D) genes and 6 Joining (J) genes.[3] The light chains also possess numerous V and J genes, but do not have D genes. By the mechanism of DNA rearrangement of these regional genes it is possible to generate an enormous antibody repertoire - 65 V genes x 27 D genes x 6 J Genes= 10530 combinations, with additional variability through light chains.
Most T cell receptors are composed of an alpha chain and a beta chain. The T cell receptor genes are similar to immunoglobulin genes in that they too contain multiple V, D and J genes in their beta chains (and V and J genes in their alpha chains) that are rearranged during the development of the lymphocyte to provide that cell with a unique antigen receptor.
V(D)J recombination of immunoglobulins
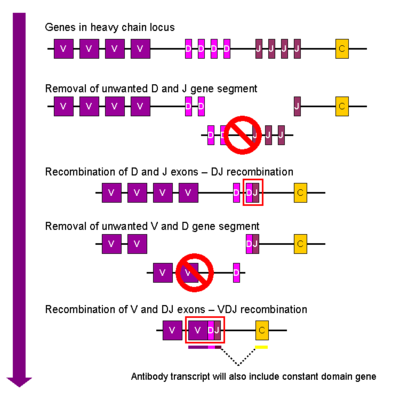
In the developing B cell, the first recombination event to occur is between one D and one J gene segment of the heavy chain locus. Any DNA between these two genes is deleted. This D-J recombination is followed by the joining of one V gene, from a region upstream of the newly formed DJ complex, forming a rearranged VDJ gene. All other genes between V and D segments of the new VDJ gene are now deleted from the cell’s genome. Primary transcript (unspliced RNA) is generated containing the VDJ region of the heavy chain and both the constant mu and delta chains (Cμ and Cδ). (i.e. the primary transcript contains the segments: V-D-J-Cμ-Cδ). The primary RNA is processed to add a polyadenylation (poly-A) tail after the Cμ chain and to remove sequence between the VDJ segment and this constant gene segment. Translation of this mRNA leads to the production of the Ig μ heavy chain protein.
The kappa (κ) and lambda (λ) chains of the immunoglobulin light chain loci rearrange in a very similar way, except the light chains lack a D segment. In other words, the first step of recombination for the light chains involves the joining of the V and J chains to give a VJ complex before the addition of the constant chain gene during primary transcription. Translation of the spliced mRNA for either the kappa or lambda chains results in formation of the Ig κ or Ig λ light chain protein.
Assembly of the Ig μ heavy chain and one of the light chains results in the formation of membrane bound form of the immunoglobulin IgM that is expressed on the surface of the immature B cell.
V(D)J recombination of the T cell receptor
During T cell development, the T cell receptor (TCR) chains undergo essentially the same sequence of ordered recombination events as that described for immunoglobulins. D-to-J recombination occurs first in the β chain of the TCR. This process can involve either the joining of the Dβ1 gene segment to one of six Jβ1 segments or the joining of the Dβ2 gene segment to one of six Jβ2 segments. DJ recombination is followed (as above) with Vβ-to-DβJβ rearrangements. All genes between the Vβ-Dβ-Jβ genes in the newly formed complex are deleted and the primary transcript is synthesized that incorporates the constant domain gene (Vβ-Dβ-Jβ-Cβ). mRNA transcription splices out any intervening sequence and allows translation of the full length protein for the TCR Cβ chain.
The rearrangement of the alpha (α) chain of the TCR follows β chain rearrangement, and resembles V-to-J rearrangement described for Ig light chains (see above). The assembly of the β- and α- chains results in formation of the αβ-TCR that is expressed on a majority of T cells.
Mechanism of recombination
Recombination Signal Sequences
The regional genes (V, D, J) are flanked by Recombination Signal Sequences (RSSs) that are recognized by a group of enzymes known collectively as the VDJ recombinase. RSSs are composed of seven conserved nucleotides (a heptamer) that reside next to the gene encoding sequence followed by a spacer (containing either 12 or 23 unconserved nucleotides) followed by a conserved nonamer (9 base pairs). The RSSs are present on the 3’ side (downstream) of a V region and the 5’ side (upstream) of the J region. These are the sides that will be involved in the joining. Only a pair of dissimilar spacer RSSs are efficiently recombined (i.e. one with a spacer of 12 nucleotides will be recombined with one that has a spacer containing 23 nucleotides). This is known as the 12/23 rule of recombination.
VDJ recombinase
VDJ recombinase refers to a collection of enzymes some of which are lymphocyte specific, and some that are expressed in many cell types. The initial steps of VDJ recombination are carried out by critical lymphocyte specific enzymes, called recombination activating gene-1 and -2 (RAG1 and RAG2). These enzymes associate with each other to recognize the RSS sequences and induce DNA cleavage at the RSS sequences. This cleavage only takes place on one strand of DNA, which leads to a nucleotide attack and creation of a hairpin loop.
Other enzymes of the VDJ recombinase are expressed in multiple cell types and are involved in DNA repair following the activity of RAG1 and RAG2. One of these enzymes is called the DNA-dependent protein kinase complex (DNA-PK) that repairs double-stranded DNA. DNA-PK binds to each end of the broken DNA and recruits several other proteins, including Artemis nuclease, XRCC4 (X-ray repair cross-complementing factor 4), DNA ligase IV, Cernunnos (also called XLF or XRCC4-like factor), and any of several DNA polymerases. DNA-PK complexes on each DNA end phosphorylate (add phosphate groups to) each other, resulting in activation of Artemis. Artemis then breaks the hairpin loop that was formed by the RAG proteins[4]. XRCC4 and Cernunnos act in concert with DNA-PK to align the two DNA ends with each other, and also help to recruit terminal transferase, which adds nucleotides randomly to the ends. DNA polymerases λ and μ insert additional nucleotides as needed to make the two ends compatible for joining. Ligase IV finally links DNA strands on opposite ends of the break to each other, completing the joining process. [5]
Because of the variability in the exact position of cleavage of the hairpin loop by Artemis, as well as the random nucleotide addition by terminal transferase, the final DNA sequence, and thus the sequence of the resulting antibody, is highly variable, even when the same two V, D, or J segments are joined. This great diversity allows VDJ recombination to generate antibodies even to microbes that neither the organism nor its ancestors have ever previously encountered.
References
- ^ Janeway CA, Jr.; et al. (2005). Immunobiology (6th ed. ed.). Garland Science. ISBN 0-443-07310-4.
{{cite book}}:|edition=has extra text (help); Explicit use of et al. in:|author=(help) - ^ Abbas AK and Lichtman AH (2003). Cellular and Molecular Immunology (5th ed. ed.). Saunders, Philadelphia. ISBN 0-7216-0008-5.
{{cite book}}:|edition=has extra text (help) - ^ Li A, Rue M, Zhou J; et al. (2004). "Utilization of Ig heavy chain variable, diversity, and joining gene segments in children with B-lineage acute lymphoblastic leukemia: implications for the mechanisms of VDJ recombination and for pathogenesis". Blood. 103 (12): 4602–9. doi:10.1182/blood-2003-11-3857. PMID 15010366.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Y. Ma, U. Pannicke, K. Schwarz and M.R. Lieber (2004). "Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination". Cell. 108: 781–794. doi:10.1016/S0092-8674(02)00671-2. PMID 11955432.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ D.C. van Gent and M. van der Burg (2007). "Non-homologous end-joining, a sticky affair". Oncogene. 26: 7731–7740. doi:10.1038/sj.onc.1210871. PMID 18066085.
Further reading
- Hartwell LH, Hood L, Goldberg ML, Reynolds AE, Silver LM, Veres RC (2000). Chapter 24, Evolution at the molecular level. In: Genetics. New York: McGraw-Hill. pp. 805–807. ISBN 0072995874.
{{cite book}}: CS1 maint: multiple names: authors list (link)
