Coronavirus nucleocapsid protein: Difference between revisions
→References: cats |
m Open access bot: pmc added to citation with #oabot. |
||
| Line 22: | Line 22: | ||
}} |
}} |
||
The '''nucleocapsid (N) protein''' is a [[protein]] that packages the [[positive-sense RNA]] [[genome]] of [[coronavirus]]es to form [[ribonucleoprotein]] structures enclosed within the viral [[capsid]].<ref name="chang_2014">{{cite journal |last1=Chang |first1=Chung-ke |last2=Hou |first2=Ming-Hon |last3=Chang |first3=Chi-Fon |last4=Hsiao |first4=Chwan-Deng |last5=Huang |first5=Tai-huang |title=The SARS coronavirus nucleocapsid protein – Forms and functions |journal=Antiviral Research |date=March 2014 |volume=103 |pages=39–50 |doi=10.1016/j.antiviral.2013.12.009}}</ref><ref name="mcbride_2014">{{cite journal |last1=McBride |first1=Ruth |last2=van Zyl |first2=Marjorie |last3=Fielding |first3=Burtram |title=The Coronavirus Nucleocapsid Is a Multifunctional Protein |journal=Viruses |date=7 August 2014 |volume=6 |issue=8 |pages=2991–3018 |doi=10.3390/v6082991}}</ref> The N protein is the most highly [[gene expression|expressed]] of the four major coronavirus [[structural protein]]s.<ref name=chang_2014 /> In addition to its interactions with [[RNA]], N forms [[protein-protein interaction]]s with the [[coronavirus membrane protein]] (M) during the process of viral assembly.<ref name=chang_2014 /><ref name=mcbride_2014 /> N also has additional functions in manipulating the [[cell cycle]] of the host cell.<ref name=mcbride_2014 /><ref name="su_2020">{{cite journal |last1=Su |first1=Mingjun |last2=Chen |first2=Yaping |last3=Qi |first3=Shanshan |last4=Shi |first4=Da |last5=Feng |first5=Li |last6=Sun |first6=Dongbo |title=A Mini-Review on Cell Cycle Regulation of Coronavirus Infection |journal=Frontiers in Veterinary Science |date=5 November 2020 |volume=7 |pages=586826 |doi=10.3389/fvets.2020.586826}}</ref> The N protein is highly [[immunogenic]] and [[antibodies]] to N are found in patients recovered from [[SARS]] and [[Covid-19]].<ref name="li_2021">{{cite journal |last1=Li |first1=Dandan |last2=Li |first2=Jinming |title=Immunologic Testing for SARS-CoV-2 Infection from the Antigen Perspective |journal=Journal of Clinical Microbiology |date=20 April 2021 |volume=59 |issue=5 |doi=10.1128/JCM.02160-20}}</ref> |
The '''nucleocapsid (N) protein''' is a [[protein]] that packages the [[positive-sense RNA]] [[genome]] of [[coronavirus]]es to form [[ribonucleoprotein]] structures enclosed within the viral [[capsid]].<ref name="chang_2014">{{cite journal |last1=Chang |first1=Chung-ke |last2=Hou |first2=Ming-Hon |last3=Chang |first3=Chi-Fon |last4=Hsiao |first4=Chwan-Deng |last5=Huang |first5=Tai-huang |title=The SARS coronavirus nucleocapsid protein – Forms and functions |journal=Antiviral Research |date=March 2014 |volume=103 |pages=39–50 |doi=10.1016/j.antiviral.2013.12.009|pmc=7113676 }}</ref><ref name="mcbride_2014">{{cite journal |last1=McBride |first1=Ruth |last2=van Zyl |first2=Marjorie |last3=Fielding |first3=Burtram |title=The Coronavirus Nucleocapsid Is a Multifunctional Protein |journal=Viruses |date=7 August 2014 |volume=6 |issue=8 |pages=2991–3018 |doi=10.3390/v6082991|pmc=4147684 }}</ref> The N protein is the most highly [[gene expression|expressed]] of the four major coronavirus [[structural protein]]s.<ref name=chang_2014 /> In addition to its interactions with [[RNA]], N forms [[protein-protein interaction]]s with the [[coronavirus membrane protein]] (M) during the process of viral assembly.<ref name=chang_2014 /><ref name=mcbride_2014 /> N also has additional functions in manipulating the [[cell cycle]] of the host cell.<ref name=mcbride_2014 /><ref name="su_2020">{{cite journal |last1=Su |first1=Mingjun |last2=Chen |first2=Yaping |last3=Qi |first3=Shanshan |last4=Shi |first4=Da |last5=Feng |first5=Li |last6=Sun |first6=Dongbo |title=A Mini-Review on Cell Cycle Regulation of Coronavirus Infection |journal=Frontiers in Veterinary Science |date=5 November 2020 |volume=7 |pages=586826 |doi=10.3389/fvets.2020.586826|pmc=7674852 }}</ref> The N protein is highly [[immunogenic]] and [[antibodies]] to N are found in patients recovered from [[SARS]] and [[Covid-19]].<ref name="li_2021">{{cite journal |last1=Li |first1=Dandan |last2=Li |first2=Jinming |title=Immunologic Testing for SARS-CoV-2 Infection from the Antigen Perspective |journal=Journal of Clinical Microbiology |date=20 April 2021 |volume=59 |issue=5 |doi=10.1128/JCM.02160-20|pmc=8091849 }}</ref> |
||
==Structure== |
==Structure== |
||
[[File:6wzo_chainAB.png|thumb|right|[[X-ray crystallography]] structure of the dimer formed by two C-terminal domains from the [[SARS-CoV-2]] N protein.<ref name=ye_2020 />]] |
[[File:6wzo_chainAB.png|thumb|right|[[X-ray crystallography]] structure of the dimer formed by two C-terminal domains from the [[SARS-CoV-2]] N protein.<ref name=ye_2020 />]] |
||
The N protein is composed of two main [[protein domain]]s connected by an [[intrinsically disordered protein|intrinsically disordered region]] (IDR) known as the linker region, with additional disordered segments at each terminus.<ref name=chang_2014 /><ref name=mcbride_2014 /> A third small domain at the C-terminal tail appears to have an ordered [[alpha helical]] [[secondary structure]] and may be involved in the formation of higher-order [[oligomer]]ic assemblies.<ref name=ye_2020 /> In [[SARS-CoV]], the causative agent of [[SARS]], the N protein is 422 [[amino acid residue]]s long<ref name=chang_2014 /> and in [[SARS-CoV-2]], the causative agent of [[Covid-19]], it is 419 residues long.<ref name="ye_2020">{{cite journal |last1=Ye |first1=Qiaozhen |last2=West |first2=Alan M. V. |last3=Silletti |first3=Steve |last4=Corbett |first4=Kevin D. |title=Architecture and self‐assembly of the SARS‐CoV ‐2 nucleocapsid protein |journal=Protein Science |date=September 2020 |volume=29 |issue=9 |pages=1890–1901 |doi=10.1002/pro.3909}}</ref><ref name="shah_2020">{{cite journal |last1=Shah |first1=Vibhuti Kumar |last2=Firmal |first2=Priyanka |last3=Alam |first3=Aftab |last4=Ganguly |first4=Dipyaman |last5=Chattopadhyay |first5=Samit |title=Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past |journal=Frontiers in Immunology |date=7 August 2020 |volume=11 |pages=1949 |doi=10.3389/fimmu.2020.01949}}</ref> |
The N protein is composed of two main [[protein domain]]s connected by an [[intrinsically disordered protein|intrinsically disordered region]] (IDR) known as the linker region, with additional disordered segments at each terminus.<ref name=chang_2014 /><ref name=mcbride_2014 /> A third small domain at the C-terminal tail appears to have an ordered [[alpha helical]] [[secondary structure]] and may be involved in the formation of higher-order [[oligomer]]ic assemblies.<ref name=ye_2020 /> In [[SARS-CoV]], the causative agent of [[SARS]], the N protein is 422 [[amino acid residue]]s long<ref name=chang_2014 /> and in [[SARS-CoV-2]], the causative agent of [[Covid-19]], it is 419 residues long.<ref name="ye_2020">{{cite journal |last1=Ye |first1=Qiaozhen |last2=West |first2=Alan M. V. |last3=Silletti |first3=Steve |last4=Corbett |first4=Kevin D. |title=Architecture and self‐assembly of the SARS‐CoV ‐2 nucleocapsid protein |journal=Protein Science |date=September 2020 |volume=29 |issue=9 |pages=1890–1901 |doi=10.1002/pro.3909}}</ref><ref name="shah_2020">{{cite journal |last1=Shah |first1=Vibhuti Kumar |last2=Firmal |first2=Priyanka |last3=Alam |first3=Aftab |last4=Ganguly |first4=Dipyaman |last5=Chattopadhyay |first5=Samit |title=Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past |journal=Frontiers in Immunology |date=7 August 2020 |volume=11 |pages=1949 |doi=10.3389/fimmu.2020.01949|pmc=7426442 }}</ref> |
||
Both the [[N-terminal]] and [[C-terminal]] domains are capable of binding [[RNA]]. The C-terminal domain forms a [[dimer]] that is likely to be the native functional state.<ref name=chang_2014 /> Parts of the IDR, particularly a [[sequence conservation|conserved]] [[sequence motif]] rich in [[serine]] and [[arginine]] residues (the SR-rich region), may also be implicated in dimer formation, though reports on this vary.<ref name=chang_2014 /><ref name=mcbride_2014 /> Although higher-order [[oligomer]]s formed through the C-terminal domain have been observed crystallographically, it is unclear if these structures have a physiological role.<ref name=chang_2014 /><ref name="chang_2016">{{cite journal |last1=Chang |first1=Chung-ke |last2=Lo |first2=Shou-Chen |last3=Wang |first3=Yong-Sheng |last4=Hou |first4=Ming-Hon |title=Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein |journal=Drug Discovery Today |date=April 2016 |volume=21 |issue=4 |pages=562–572 |doi=10.1016/j.drudis.2015.11.015}}</ref> |
Both the [[N-terminal]] and [[C-terminal]] domains are capable of binding [[RNA]]. The C-terminal domain forms a [[dimer]] that is likely to be the native functional state.<ref name=chang_2014 /> Parts of the IDR, particularly a [[sequence conservation|conserved]] [[sequence motif]] rich in [[serine]] and [[arginine]] residues (the SR-rich region), may also be implicated in dimer formation, though reports on this vary.<ref name=chang_2014 /><ref name=mcbride_2014 /> Although higher-order [[oligomer]]s formed through the C-terminal domain have been observed crystallographically, it is unclear if these structures have a physiological role.<ref name=chang_2014 /><ref name="chang_2016">{{cite journal |last1=Chang |first1=Chung-ke |last2=Lo |first2=Shou-Chen |last3=Wang |first3=Yong-Sheng |last4=Hou |first4=Ming-Hon |title=Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein |journal=Drug Discovery Today |date=April 2016 |volume=21 |issue=4 |pages=562–572 |doi=10.1016/j.drudis.2015.11.015|pmc=7108309 }}</ref> |
||
The C-terminal dimer has been structurally characterized by [[X-ray crystallography]] for several coronaviruses and has a highly conserved structure.<ref name=ye_2020 /> The N-terminal domain - sometimes known as the RNA-binding domain, though other parts of the protein also interact with RNA - has also been crystallized and has been studied by [[protein NMR|nuclear magnetic resonance spectroscopy]] in the presence of RNA.<ref name="dinesh_2020">{{cite journal |last1=Dinesh |first1=Dhurvas Chandrasekaran |last2=Chalupska |first2=Dominika |last3=Silhan |first3=Jan |last4=Koutna |first4=Eliska |last5=Nencka |first5=Radim |last6=Veverka |first6=Vaclav |last7=Boura |first7=Evzen |title=Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein |journal=PLOS Pathogens |date=2 December 2020 |volume=16 |issue=12 |pages=e1009100 |doi=10.1371/journal.ppat.1009100}}</ref> |
The C-terminal dimer has been structurally characterized by [[X-ray crystallography]] for several coronaviruses and has a highly conserved structure.<ref name=ye_2020 /> The N-terminal domain - sometimes known as the RNA-binding domain, though other parts of the protein also interact with RNA - has also been crystallized and has been studied by [[protein NMR|nuclear magnetic resonance spectroscopy]] in the presence of RNA.<ref name="dinesh_2020">{{cite journal |last1=Dinesh |first1=Dhurvas Chandrasekaran |last2=Chalupska |first2=Dominika |last3=Silhan |first3=Jan |last4=Koutna |first4=Eliska |last5=Nencka |first5=Radim |last6=Veverka |first6=Vaclav |last7=Boura |first7=Evzen |title=Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein |journal=PLOS Pathogens |date=2 December 2020 |volume=16 |issue=12 |pages=e1009100 |doi=10.1371/journal.ppat.1009100|pmc=7735635 }}</ref> |
||
===Post-translational modifications=== |
===Post-translational modifications=== |
||
The N protein is [[post-translational modification|post-translationally modified]] by [[phosphorylation]] at sites located in the IDR, particularly in the SR-rich region.<ref name=chang_2014 /><ref name="fung_2018">{{cite journal |last1=Fung |first1=To Sing |last2=Liu |first2=Ding Xiang |title=Post-translational modifications of coronavirus proteins: roles and function |journal=Future Virology |date=June 2018 |volume=13 |issue=6 |pages=405–430 |doi=10.2217/fvl-2018-0008}}</ref> In several coronaviruses, [[ADP-ribosylation]] of the N protein has also been reported.<ref name="grunewald_2018">{{cite journal |last1=Grunewald |first1=Matthew E. |last2=Fehr |first2=Anthony R. |last3=Athmer |first3=Jeremiah |last4=Perlman |first4=Stanley |title=The coronavirus nucleocapsid protein is ADP-ribosylated |journal=Virology |date=April 2018 |volume=517 |pages=62–68 |doi=10.1016/j.virol.2017.11.020}}</ref><ref name=fung_2018 /> With unclear functional significance, the SARS-CoV N protein has been observed to be [[SUMOylated]] and the N proteins of several coronaviruses have been observed to be [[proteolysis|proteolytically cleaved]].<ref name=fung_2018 /> |
The N protein is [[post-translational modification|post-translationally modified]] by [[phosphorylation]] at sites located in the IDR, particularly in the SR-rich region.<ref name=chang_2014 /><ref name="fung_2018">{{cite journal |last1=Fung |first1=To Sing |last2=Liu |first2=Ding Xiang |title=Post-translational modifications of coronavirus proteins: roles and function |journal=Future Virology |date=June 2018 |volume=13 |issue=6 |pages=405–430 |doi=10.2217/fvl-2018-0008|pmc=7080180 }}</ref> In several coronaviruses, [[ADP-ribosylation]] of the N protein has also been reported.<ref name="grunewald_2018">{{cite journal |last1=Grunewald |first1=Matthew E. |last2=Fehr |first2=Anthony R. |last3=Athmer |first3=Jeremiah |last4=Perlman |first4=Stanley |title=The coronavirus nucleocapsid protein is ADP-ribosylated |journal=Virology |date=April 2018 |volume=517 |pages=62–68 |doi=10.1016/j.virol.2017.11.020|pmc=5871557 }}</ref><ref name=fung_2018 /> With unclear functional significance, the SARS-CoV N protein has been observed to be [[SUMOylated]] and the N proteins of several coronaviruses have been observed to be [[proteolysis|proteolytically cleaved]].<ref name=fung_2018 /> |
||
==Expression and localization== |
==Expression and localization== |
||
The N protein is the most highly [[gene expression|expressed]] in host cells of the four major [[structural protein]]s.<ref name=chang_2014 /> Like the other structural proteins, the [[gene]] encoding the N protein is located toward the [[3' end]] of the [[genome]].<ref name=mcbride_2014 /> |
The N protein is the most highly [[gene expression|expressed]] in host cells of the four major [[structural protein]]s.<ref name=chang_2014 /> Like the other structural proteins, the [[gene]] encoding the N protein is located toward the [[3' end]] of the [[genome]].<ref name=mcbride_2014 /> |
||
N protein is [[subcellular localization|localized]] primarily to the [[cytoplasm]].<ref name=mcbride_2014 /> In many coronaviruses, a population of N protein is localized to the [[nucleolus]],<ref name=mcbride_2014 /><ref name=su_2020 /><ref name="masters_2006">{{cite journal |last1=Masters |first1=Paul S. |title=The Molecular Biology of Coronaviruses |journal=Advances in Virus Research |date=2006 |volume=66 |pages=193–292 |doi=10.1016/S0065-3527(06)66005-3}}</ref> thought to be associated with its effects on the [[cell cycle]].<ref name=su_2020 /> |
N protein is [[subcellular localization|localized]] primarily to the [[cytoplasm]].<ref name=mcbride_2014 /> In many coronaviruses, a population of N protein is localized to the [[nucleolus]],<ref name=mcbride_2014 /><ref name=su_2020 /><ref name="masters_2006">{{cite journal |last1=Masters |first1=Paul S. |title=The Molecular Biology of Coronaviruses |journal=Advances in Virus Research |date=2006 |volume=66 |pages=193–292 |doi=10.1016/S0065-3527(06)66005-3|pmc=7112330 }}</ref> thought to be associated with its effects on the [[cell cycle]].<ref name=su_2020 /> |
||
==Function== |
==Function== |
||
| Line 47: | Line 47: | ||
===Genomic and subgenomic RNA synthesis=== |
===Genomic and subgenomic RNA synthesis=== |
||
Synthesis of genomic RNA appears to involve participation by the N protein. N is physically colocalized with the viral [[RNA-dependent RNA polymerase]] early in the replication cycle and forms interactions with [[coronavirus non-structural protein 3|non-structural protein 3]], a component of the [[Coronavirus#Replicase-transcriptase|replicase-transcriptase complex]].<ref name=mcbride_2014 /> Although N appears to facilitate efficient replication of genomic RNA, it is not required for RNA transcription in all coronaviruses.<ref name=mcbride_2014 /><ref name="zuniga_2010">{{cite journal |last1=Zúñiga |first1=Sonia |last2=Cruz |first2=Jazmina L. G. |last3=Sola |first3=Isabel |last4=Mateos-Gómez |first4=Pedro A. |last5=Palacio |first5=Lorena |last6=Enjuanes |first6=Luis |title=Coronavirus Nucleocapsid Protein Facilitates Template Switching and Is Required for Efficient Transcription |journal=Journal of Virology |date=15 February 2010 |volume=84 |issue=4 |pages=2169–2175 |doi=10.1128/JVI.02011-09}}</ref> In at least one coronavirus, [[transmissible gastroenteritis virus]] (TGEV), N is involved in template switching in the production of [[subgenomic mRNA]]s, a process that is a distinctive feature of viruses in the order ''[[Nidovirales]]''.<ref name=mcbride_2014 /><ref name=zuniga_2010 /><ref name="sola_2015">{{cite journal |last1=Sola |first1=Isabel |last2=Almazán |first2=Fernando |last3=Zúñiga |first3=Sonia |last4=Enjuanes |first4=Luis |title=Continuous and Discontinuous RNA Synthesis in Coronaviruses |journal=Annual Review of Virology |date=9 November 2015 |volume=2 |issue=1 |pages=265–288 |doi=10.1146/annurev-virology-100114-055218}}</ref> |
Synthesis of genomic RNA appears to involve participation by the N protein. N is physically colocalized with the viral [[RNA-dependent RNA polymerase]] early in the replication cycle and forms interactions with [[coronavirus non-structural protein 3|non-structural protein 3]], a component of the [[Coronavirus#Replicase-transcriptase|replicase-transcriptase complex]].<ref name=mcbride_2014 /> Although N appears to facilitate efficient replication of genomic RNA, it is not required for RNA transcription in all coronaviruses.<ref name=mcbride_2014 /><ref name="zuniga_2010">{{cite journal |last1=Zúñiga |first1=Sonia |last2=Cruz |first2=Jazmina L. G. |last3=Sola |first3=Isabel |last4=Mateos-Gómez |first4=Pedro A. |last5=Palacio |first5=Lorena |last6=Enjuanes |first6=Luis |title=Coronavirus Nucleocapsid Protein Facilitates Template Switching and Is Required for Efficient Transcription |journal=Journal of Virology |date=15 February 2010 |volume=84 |issue=4 |pages=2169–2175 |doi=10.1128/JVI.02011-09|pmc=2812394 }}</ref> In at least one coronavirus, [[transmissible gastroenteritis virus]] (TGEV), N is involved in template switching in the production of [[subgenomic mRNA]]s, a process that is a distinctive feature of viruses in the order ''[[Nidovirales]]''.<ref name=mcbride_2014 /><ref name=zuniga_2010 /><ref name="sola_2015">{{cite journal |last1=Sola |first1=Isabel |last2=Almazán |first2=Fernando |last3=Zúñiga |first3=Sonia |last4=Enjuanes |first4=Luis |title=Continuous and Discontinuous RNA Synthesis in Coronaviruses |journal=Annual Review of Virology |date=9 November 2015 |volume=2 |issue=1 |pages=265–288 |doi=10.1146/annurev-virology-100114-055218|pmc=6025776 }}</ref> |
||
===Cell cycle effects=== |
===Cell cycle effects=== |
||
| Line 53: | Line 53: | ||
===Immune system effects=== |
===Immune system effects=== |
||
The N protein is involved in [[viral pathogenesis]] via its effects on components of the [[immune system]]. In [[SARS-CoV]],<ref name=mcbride_2014 /><ref name="speigel_2005">{{cite journal |last1=Spiegel |first1=Martin |last2=Pichlmair |first2=Andreas |last3=Martínez-Sobrido |first3=Luis |last4=Cros |first4=Jerome |last5=García-Sastre |first5=Adolfo |last6=Haller |first6=Otto |last7=Weber |first7=Friedemann |title=Inhibition of Beta Interferon Induction by Severe Acute Respiratory Syndrome Coronavirus Suggests a Two-Step Model for Activation of Interferon Regulatory Factor 3 |journal=Journal of Virology |date=15 February 2005 |volume=79 |issue=4 |pages=2079–2086 |doi=10.1128/JVI.79.4.2079-2086.2005}}</ref><ref name="kopecky_2007">{{cite journal |last1=Kopecky-Bromberg |first1=Sarah A. |last2=Martínez-Sobrido |first2=Luis |last3=Frieman |first3=Matthew |last4=Baric |first4=Ralph A. |last5=Palese |first5=Peter |title=Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists |journal=Journal of Virology |date=15 January 2007 |volume=81 |issue=2 |pages=548–557 |doi=10.1128/JVI.01782-06}}</ref> [[MERS-CoV]],<ref name="chang_2020">{{cite journal |last1=Chang |first1=Chi-You |last2=Liu |first2=Helene Minyi |last3=Chang |first3=Ming-Fu |last4=Chang |first4=Shin C. |title=Middle East Respiratory Syndrome Coronavirus Nucleocapsid Protein Suppresses Type I and Type III Interferon Induction by Targeting RIG-I Signaling |journal=Journal of Virology |date=16 June 2020 |volume=94 |issue=13 |doi=10.1128/JVI.00099-20}}</ref> and [[SARS-CoV-2]],<ref name="mu_2020">{{cite journal |last1=Mu |first1=Jingfang |last2=Fang |first2=Yaohui |last3=Yang |first3=Qi |last4=Shu |first4=Ting |last5=Wang |first5=An |last6=Huang |first6=Muhan |last7=Jin |first7=Liang |last8=Deng |first8=Fei |last9=Qiu |first9=Yang |last10=Zhou |first10=Xi |title=SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2 |journal=Cell Discovery |date=December 2020 |volume=6 |issue=1 |pages=65 |doi=10.1038/s41421-020-00208-3}}</ref> N has been reported as suppressing [[interferon]] responses. |
The N protein is involved in [[viral pathogenesis]] via its effects on components of the [[immune system]]. In [[SARS-CoV]],<ref name=mcbride_2014 /><ref name="speigel_2005">{{cite journal |last1=Spiegel |first1=Martin |last2=Pichlmair |first2=Andreas |last3=Martínez-Sobrido |first3=Luis |last4=Cros |first4=Jerome |last5=García-Sastre |first5=Adolfo |last6=Haller |first6=Otto |last7=Weber |first7=Friedemann |title=Inhibition of Beta Interferon Induction by Severe Acute Respiratory Syndrome Coronavirus Suggests a Two-Step Model for Activation of Interferon Regulatory Factor 3 |journal=Journal of Virology |date=15 February 2005 |volume=79 |issue=4 |pages=2079–2086 |doi=10.1128/JVI.79.4.2079-2086.2005|pmc=546554 }}</ref><ref name="kopecky_2007">{{cite journal |last1=Kopecky-Bromberg |first1=Sarah A. |last2=Martínez-Sobrido |first2=Luis |last3=Frieman |first3=Matthew |last4=Baric |first4=Ralph A. |last5=Palese |first5=Peter |title=Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists |journal=Journal of Virology |date=15 January 2007 |volume=81 |issue=2 |pages=548–557 |doi=10.1128/JVI.01782-06|pmc=1797484 }}</ref> [[MERS-CoV]],<ref name="chang_2020">{{cite journal |last1=Chang |first1=Chi-You |last2=Liu |first2=Helene Minyi |last3=Chang |first3=Ming-Fu |last4=Chang |first4=Shin C. |title=Middle East Respiratory Syndrome Coronavirus Nucleocapsid Protein Suppresses Type I and Type III Interferon Induction by Targeting RIG-I Signaling |journal=Journal of Virology |date=16 June 2020 |volume=94 |issue=13 |doi=10.1128/JVI.00099-20|pmc=7307178 }}</ref> and [[SARS-CoV-2]],<ref name="mu_2020">{{cite journal |last1=Mu |first1=Jingfang |last2=Fang |first2=Yaohui |last3=Yang |first3=Qi |last4=Shu |first4=Ting |last5=Wang |first5=An |last6=Huang |first6=Muhan |last7=Jin |first7=Liang |last8=Deng |first8=Fei |last9=Qiu |first9=Yang |last10=Zhou |first10=Xi |title=SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2 |journal=Cell Discovery |date=December 2020 |volume=6 |issue=1 |pages=65 |doi=10.1038/s41421-020-00208-3|pmc=7490572 }}</ref> N has been reported as suppressing [[interferon]] responses. |
||
==Evolution and conservation== |
==Evolution and conservation== |
||
The structures of N proteins from different coronaviruses, particularly the C-terminal domains, appear to be well conserved.<ref name=chang_2014 /><ref name=ye_2020 /> Similarities between the structure and topology of the N proteins of coronaviruses and [[arterivirus]]es suggest a common evolutionary origin and supports the classification of these two groups in the common order ''[[Nidovirales]]''.<ref name=chang_2014 /><ref name=mcbride_2014 /> |
The structures of N proteins from different coronaviruses, particularly the C-terminal domains, appear to be well conserved.<ref name=chang_2014 /><ref name=ye_2020 /> Similarities between the structure and topology of the N proteins of coronaviruses and [[arterivirus]]es suggest a common evolutionary origin and supports the classification of these two groups in the common order ''[[Nidovirales]]''.<ref name=chang_2014 /><ref name=mcbride_2014 /> |
||
Examination of SARS-CoV-2 sequences collected during the [[Covid-19 pandemic]] found that [[missense mutation]]s were most common in the central linker region of the protein, suggesting this relatively unstructured region is more tolerant of mutations than the structured domains.<ref name=ye_2020 /> A separate study of SARS-CoV-2 sequences identified at least one site in the N protein under [[positive selection]].<ref name="cagliani_2020">{{cite journal |last1=Cagliani |first1=Rachele |last2=Forni |first2=Diego |last3=Clerici |first3=Mario |last4=Sironi |first4=Manuela |title=Computational Inference of Selection Underlying the Evolution of the Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 |journal=Journal of Virology |date=June 2020 |volume=94 |issue=12 |doi=10.1128/JVI.00411-20}}</ref> |
Examination of SARS-CoV-2 sequences collected during the [[Covid-19 pandemic]] found that [[missense mutation]]s were most common in the central linker region of the protein, suggesting this relatively unstructured region is more tolerant of mutations than the structured domains.<ref name=ye_2020 /> A separate study of SARS-CoV-2 sequences identified at least one site in the N protein under [[positive selection]].<ref name="cagliani_2020">{{cite journal |last1=Cagliani |first1=Rachele |last2=Forni |first2=Diego |last3=Clerici |first3=Mario |last4=Sironi |first4=Manuela |title=Computational Inference of Selection Underlying the Evolution of the Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 |journal=Journal of Virology |date=June 2020 |volume=94 |issue=12 |doi=10.1128/JVI.00411-20|pmc=7307108 }}</ref> |
||
==References== |
==References== |
||
Revision as of 19:41, 25 July 2021
| Nucleocapsid protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Illustration of a SARS-CoV-2 virion.[1] The N protein, contained entirely within the virion, is not visible. Red: spike proteins (S) Yellow: envelope proteins (E) Orange: membrane proteins (M) | |||||||||
| Identifiers | |||||||||
| Symbol | CoV_nucleocap | ||||||||
| Pfam | PF00937 | ||||||||
| InterPro | IPR001218 | ||||||||
| |||||||||
The nucleocapsid (N) protein is a protein that packages the positive-sense RNA genome of coronaviruses to form ribonucleoprotein structures enclosed within the viral capsid.[2][3] The N protein is the most highly expressed of the four major coronavirus structural proteins.[2] In addition to its interactions with RNA, N forms protein-protein interactions with the coronavirus membrane protein (M) during the process of viral assembly.[2][3] N also has additional functions in manipulating the cell cycle of the host cell.[3][4] The N protein is highly immunogenic and antibodies to N are found in patients recovered from SARS and Covid-19.[5]
Structure
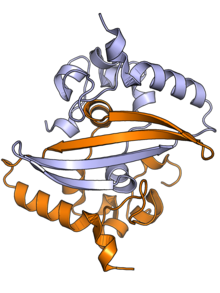
The N protein is composed of two main protein domains connected by an intrinsically disordered region (IDR) known as the linker region, with additional disordered segments at each terminus.[2][3] A third small domain at the C-terminal tail appears to have an ordered alpha helical secondary structure and may be involved in the formation of higher-order oligomeric assemblies.[6] In SARS-CoV, the causative agent of SARS, the N protein is 422 amino acid residues long[2] and in SARS-CoV-2, the causative agent of Covid-19, it is 419 residues long.[6][7]
Both the N-terminal and C-terminal domains are capable of binding RNA. The C-terminal domain forms a dimer that is likely to be the native functional state.[2] Parts of the IDR, particularly a conserved sequence motif rich in serine and arginine residues (the SR-rich region), may also be implicated in dimer formation, though reports on this vary.[2][3] Although higher-order oligomers formed through the C-terminal domain have been observed crystallographically, it is unclear if these structures have a physiological role.[2][8]
The C-terminal dimer has been structurally characterized by X-ray crystallography for several coronaviruses and has a highly conserved structure.[6] The N-terminal domain - sometimes known as the RNA-binding domain, though other parts of the protein also interact with RNA - has also been crystallized and has been studied by nuclear magnetic resonance spectroscopy in the presence of RNA.[9]
Post-translational modifications
The N protein is post-translationally modified by phosphorylation at sites located in the IDR, particularly in the SR-rich region.[2][10] In several coronaviruses, ADP-ribosylation of the N protein has also been reported.[11][10] With unclear functional significance, the SARS-CoV N protein has been observed to be SUMOylated and the N proteins of several coronaviruses have been observed to be proteolytically cleaved.[10]
Expression and localization
The N protein is the most highly expressed in host cells of the four major structural proteins.[2] Like the other structural proteins, the gene encoding the N protein is located toward the 3' end of the genome.[3]
N protein is localized primarily to the cytoplasm.[3] In many coronaviruses, a population of N protein is localized to the nucleolus,[3][4][12] thought to be associated with its effects on the cell cycle.[4]
Function
Genome packaging and viral assembly

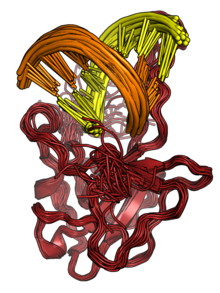
The N protein binds to RNA to form ribonucleoprotein (RNP) structures for packaging the genome into the viral capsid.[2][3] The RNP particles formed are roughly spherical and are organized in flexible helical structures inside the virus.[2][3] Formation of RNPs is thought to involve allosteric interactions between RNA and multiple RNA-binding regions of the protein.[2][8] Dimerization of N is important for assembly of RNPs. Encapsidation of the genome occurs through interactions between N and M.[2][3] N is essential for viral assembly.[3] N also serves as a chaperone protein for the formation of RNA structure in the genomic RNA.[3][8]
Genomic and subgenomic RNA synthesis
Synthesis of genomic RNA appears to involve participation by the N protein. N is physically colocalized with the viral RNA-dependent RNA polymerase early in the replication cycle and forms interactions with non-structural protein 3, a component of the replicase-transcriptase complex.[3] Although N appears to facilitate efficient replication of genomic RNA, it is not required for RNA transcription in all coronaviruses.[3][13] In at least one coronavirus, transmissible gastroenteritis virus (TGEV), N is involved in template switching in the production of subgenomic mRNAs, a process that is a distinctive feature of viruses in the order Nidovirales.[3][13][14]
Cell cycle effects
Coronaviruses manipulate the cell cycle of the host cell through various mechanisms. In several coronaviruses, including SARS-CoV, the N protein has been reported to cause cell cycle arrest in S phase through interactions with cyclin-CDK.[3][4] In SARS-CoV, a cyclin box-binding region in the N protein can serve as a cyclin-CDK phosphorylation substrate.[3] Trafficking of N to the nucleolus may also play a role in cell cycle effects.[4] More broadly, N may be involved in reduction of host cell protein translation activity.[3]
Immune system effects
The N protein is involved in viral pathogenesis via its effects on components of the immune system. In SARS-CoV,[3][15][16] MERS-CoV,[17] and SARS-CoV-2,[18] N has been reported as suppressing interferon responses.
Evolution and conservation
The structures of N proteins from different coronaviruses, particularly the C-terminal domains, appear to be well conserved.[2][6] Similarities between the structure and topology of the N proteins of coronaviruses and arteriviruses suggest a common evolutionary origin and supports the classification of these two groups in the common order Nidovirales.[2][3]
Examination of SARS-CoV-2 sequences collected during the Covid-19 pandemic found that missense mutations were most common in the central linker region of the protein, suggesting this relatively unstructured region is more tolerant of mutations than the structured domains.[6] A separate study of SARS-CoV-2 sequences identified at least one site in the N protein under positive selection.[19]
References
- ^ Giaimo C (1 April 2020). "The Spiky Blob Seen Around the World". The New York Times. Archived from the original on 2 April 2020. Retrieved 6 April 2020.
- ^ a b c d e f g h i j k l m n o p Chang, Chung-ke; Hou, Ming-Hon; Chang, Chi-Fon; Hsiao, Chwan-Deng; Huang, Tai-huang (March 2014). "The SARS coronavirus nucleocapsid protein – Forms and functions". Antiviral Research. 103: 39–50. doi:10.1016/j.antiviral.2013.12.009. PMC 7113676.
- ^ a b c d e f g h i j k l m n o p q r s t u McBride, Ruth; van Zyl, Marjorie; Fielding, Burtram (7 August 2014). "The Coronavirus Nucleocapsid Is a Multifunctional Protein". Viruses. 6 (8): 2991–3018. doi:10.3390/v6082991. PMC 4147684.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e Su, Mingjun; Chen, Yaping; Qi, Shanshan; Shi, Da; Feng, Li; Sun, Dongbo (5 November 2020). "A Mini-Review on Cell Cycle Regulation of Coronavirus Infection". Frontiers in Veterinary Science. 7: 586826. doi:10.3389/fvets.2020.586826. PMC 7674852.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Li, Dandan; Li, Jinming (20 April 2021). "Immunologic Testing for SARS-CoV-2 Infection from the Antigen Perspective". Journal of Clinical Microbiology. 59 (5). doi:10.1128/JCM.02160-20. PMC 8091849.
- ^ a b c d e f Ye, Qiaozhen; West, Alan M. V.; Silletti, Steve; Corbett, Kevin D. (September 2020). "Architecture and self‐assembly of the SARS‐CoV ‐2 nucleocapsid protein". Protein Science. 29 (9): 1890–1901. doi:10.1002/pro.3909.
- ^ Shah, Vibhuti Kumar; Firmal, Priyanka; Alam, Aftab; Ganguly, Dipyaman; Chattopadhyay, Samit (7 August 2020). "Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past". Frontiers in Immunology. 11: 1949. doi:10.3389/fimmu.2020.01949. PMC 7426442.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c Chang, Chung-ke; Lo, Shou-Chen; Wang, Yong-Sheng; Hou, Ming-Hon (April 2016). "Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein". Drug Discovery Today. 21 (4): 562–572. doi:10.1016/j.drudis.2015.11.015. PMC 7108309.
- ^ a b Dinesh, Dhurvas Chandrasekaran; Chalupska, Dominika; Silhan, Jan; Koutna, Eliska; Nencka, Radim; Veverka, Vaclav; Boura, Evzen (2 December 2020). "Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein". PLOS Pathogens. 16 (12): e1009100. doi:10.1371/journal.ppat.1009100. PMC 7735635.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c Fung, To Sing; Liu, Ding Xiang (June 2018). "Post-translational modifications of coronavirus proteins: roles and function". Future Virology. 13 (6): 405–430. doi:10.2217/fvl-2018-0008. PMC 7080180.
- ^ Grunewald, Matthew E.; Fehr, Anthony R.; Athmer, Jeremiah; Perlman, Stanley (April 2018). "The coronavirus nucleocapsid protein is ADP-ribosylated". Virology. 517: 62–68. doi:10.1016/j.virol.2017.11.020. PMC 5871557.
- ^ Masters, Paul S. (2006). "The Molecular Biology of Coronaviruses". Advances in Virus Research. 66: 193–292. doi:10.1016/S0065-3527(06)66005-3. PMC 7112330.
- ^ a b Zúñiga, Sonia; Cruz, Jazmina L. G.; Sola, Isabel; Mateos-Gómez, Pedro A.; Palacio, Lorena; Enjuanes, Luis (15 February 2010). "Coronavirus Nucleocapsid Protein Facilitates Template Switching and Is Required for Efficient Transcription". Journal of Virology. 84 (4): 2169–2175. doi:10.1128/JVI.02011-09. PMC 2812394.
- ^ Sola, Isabel; Almazán, Fernando; Zúñiga, Sonia; Enjuanes, Luis (9 November 2015). "Continuous and Discontinuous RNA Synthesis in Coronaviruses". Annual Review of Virology. 2 (1): 265–288. doi:10.1146/annurev-virology-100114-055218. PMC 6025776.
- ^ Spiegel, Martin; Pichlmair, Andreas; Martínez-Sobrido, Luis; Cros, Jerome; García-Sastre, Adolfo; Haller, Otto; Weber, Friedemann (15 February 2005). "Inhibition of Beta Interferon Induction by Severe Acute Respiratory Syndrome Coronavirus Suggests a Two-Step Model for Activation of Interferon Regulatory Factor 3". Journal of Virology. 79 (4): 2079–2086. doi:10.1128/JVI.79.4.2079-2086.2005. PMC 546554.
- ^ Kopecky-Bromberg, Sarah A.; Martínez-Sobrido, Luis; Frieman, Matthew; Baric, Ralph A.; Palese, Peter (15 January 2007). "Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists". Journal of Virology. 81 (2): 548–557. doi:10.1128/JVI.01782-06. PMC 1797484.
- ^ Chang, Chi-You; Liu, Helene Minyi; Chang, Ming-Fu; Chang, Shin C. (16 June 2020). "Middle East Respiratory Syndrome Coronavirus Nucleocapsid Protein Suppresses Type I and Type III Interferon Induction by Targeting RIG-I Signaling". Journal of Virology. 94 (13). doi:10.1128/JVI.00099-20. PMC 7307178.
- ^ Mu, Jingfang; Fang, Yaohui; Yang, Qi; Shu, Ting; Wang, An; Huang, Muhan; Jin, Liang; Deng, Fei; Qiu, Yang; Zhou, Xi (December 2020). "SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2". Cell Discovery. 6 (1): 65. doi:10.1038/s41421-020-00208-3. PMC 7490572.
- ^ Cagliani, Rachele; Forni, Diego; Clerici, Mario; Sironi, Manuela (June 2020). "Computational Inference of Selection Underlying the Evolution of the Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2". Journal of Virology. 94 (12). doi:10.1128/JVI.00411-20. PMC 7307108.
