Haloquadratum walsbyi: Difference between revisions
more added to normal microbiota |
added more information to history and added a citation |
||
| Line 35: | Line 35: | ||
== Diversity == |
== Diversity == |
||
{{clear|right}}A surprisingly high amount of cells in salt brines around the world are ''Haloquadratum walsbyi'' |
{{clear|right}}A surprisingly high amount of cells in salt brines around the world are ''Haloquadratum walsbyi.'' Up to 80% of the cells in the brines can be identified as ''Haloquadratum walsbyi.'' Experiments have been done to examine the genetic diversity in the salt brine environment. Seven different types of ''H. walsbyi's'' genomic island have been discovered in natural environments.<ref name=":5">{{Cite journal |last=Martin-Cuadrado |first=Ana-Belen |last2=Pašić |first2=Lejla |last3=Rodriguez-Valera |first3=Francisco |date=2015-08-13 |title=Diversity of the cell-wall associated genomic island of the archaeon Haloquadratum walsbyi |url=https://doi.org/10.1186/s12864-015-1794-8 |journal=BMC Genomics |volume=16 |issue=1 |pages=603 |doi=10.1186/s12864-015-1794-8 |issn=1471-2164 |pmc=PMC4535781 |pmid=26268990}}</ref> After examining the [[Metagenomics|metogenomic]] [[fosmid]] library for ''H. walsbyi,'' two types of the cell-wall associated islands were identified. The genes in these islands include those responsible for the synthesis of surface layer structures such as [[Glycoprotein|glycoproteins]] and genes responsible for the synthesis of [[Cell envelope|cell envelopes]].<ref name=":4" /> [[Homologous recombination]] is responsible for the maintaining the genes mentioned above and also the diversity of the metagenome in its natural environment. Surface structures on different ''H. walsbyi'' cells help to differentiate sources of lineage for the population as a whole. These differing structures also increase the diversity of the cells in their natural environment. These changes in cell structure may be due to the cells' attempts to reduce their susceptibility to attack by [[Virus|viruses]].<ref name=":5" /> |
||
== Genomics and Structure == |
== Genomics and Structure == |
||
''H. walsbyi'' is classified as an [[oligotroph]]ic microorganism, as it grows in nutrient deficient conditions where concentrations of organic substances are minimal. To combat, ''H. walsbyi'' maintains a high [[Surface-area-to-volume ratio|surface to volume ratio]] by flattening to maximize nutrient uptake. Because of their [[Haloquadratum|square shape]], they are more capable of flattening than [[Sphere|spherical]] shaped microorganisms are.<ref name=":2" /> ''H. walsbyi'' can flatten an extreme amount of about 0.1-0.5μm. The overall size of the cell structure ranges from 1.5 to 11 μm. However, larger cells have been observed. The largest recorded H. walsbyi cell was measured as 40 x 40 μm.<ref name=":3">{{Cite journal |last=Zenke |first=Ralf |last2=von Gronau |first2=Susanne |last3=Bolhuis |first3=Henk |last4=Gruska |first4=Manuela |last5=Pfeiffer |first5=Friedhelm |last6=Oesterhelt |first6=Dieter |date=2015 |title=Fluorescence microscopy visualization of halomucin, a secreted 927 kDa protein surrounding Haloquadratum walsbyi cells |url=https://www.frontiersin.org/articles/10.3389/fmicb.2015.00249 |journal=Frontiers in Microbiology |volume=6 |doi=10.3389/fmicb.2015.00249/full |issn=1664-302X}}</ref> |
''H. walsbyi'' is classified as an [[oligotroph]]ic microorganism, as it grows in nutrient deficient conditions where concentrations of organic substances are minimal. To combat, ''H. walsbyi'' maintains a high [[Surface-area-to-volume ratio|surface to volume ratio]] by flattening to maximize nutrient uptake. Because of their [[Haloquadratum|square shape]], they are more capable of flattening than [[Sphere|spherical]] shaped microorganisms are.<ref name=":2" /> ''H. walsbyi'' can flatten an extreme amount of about 0.1-0.5μm. The overall size of the cell structure ranges from 1.5 to 11 μm. However, larger cells have been observed. The largest recorded H. walsbyi cell was measured as 40 x 40 μm.<ref name=":3">{{Cite journal |last=Zenke |first=Ralf |last2=von Gronau |first2=Susanne |last3=Bolhuis |first3=Henk |last4=Gruska |first4=Manuela |last5=Pfeiffer |first5=Friedhelm |last6=Oesterhelt |first6=Dieter |date=2015 |title=Fluorescence microscopy visualization of halomucin, a secreted 927 kDa protein surrounding Haloquadratum walsbyi cells |url=https://www.frontiersin.org/articles/10.3389/fmicb.2015.00249 |journal=Frontiers in Microbiology |volume=6 |doi=10.3389/fmicb.2015.00249/full |issn=1664-302X}}</ref> |
||
| Line 48: | Line 48: | ||
Upon the observation of the unique shape of ''H. walsbyi'', cultivation has been a goal for scientists studying the species. Hyper-saline media has been found to be a substantial medium to maintain the pure cultures.<ref name=":6">{{Cite journal |last=Bolhuis |first=Henk |last2=Poele |first2=Evelien M. te |last3=Rodriguez-Valera |first3=Francisco |date=24 September 2004 |title=Isolation and cultivation of Walsby's square archaeon |url=https://onlinelibrary.wiley.com/doi/10.1111/j.1462-2920.2004.00692.x |journal=Environmental Microbiology |language=en |volume=6 |issue=12 |pages=1287–1291 |doi=10.1111/j.1462-2920.2004.00692.x |issn=1462-2912}}</ref> ''H. walsbyi'' remains one of the largest prokaryotes known today and contains roughly 3 million basepairs.<ref name=":6" /> |
Upon the observation of the unique shape of ''H. walsbyi'', cultivation has been a goal for scientists studying the species. Hyper-saline media has been found to be a substantial medium to maintain the pure cultures.<ref name=":6">{{Cite journal |last=Bolhuis |first=Henk |last2=Poele |first2=Evelien M. te |last3=Rodriguez-Valera |first3=Francisco |date=24 September 2004 |title=Isolation and cultivation of Walsby's square archaeon |url=https://onlinelibrary.wiley.com/doi/10.1111/j.1462-2920.2004.00692.x |journal=Environmental Microbiology |language=en |volume=6 |issue=12 |pages=1287–1291 |doi=10.1111/j.1462-2920.2004.00692.x |issn=1462-2912}}</ref> ''H. walsbyi'' remains one of the largest prokaryotes known today and contains roughly 3 million basepairs.<ref name=":6" /> |
||
In 2004, two strains of ''H. walsbyi'' were successfully isolated and able to be sequenced. The first strain was an isolate from the Spanish, called HBSQ00.<ref name=":02">{{cite journal |vauthors=Bolhuis H, Martín-Cuadrado AB, Rosselli R, Pašić L, Rodriguez-Valera F |date=July 2017 |title=Transcriptome analysis of Haloquadratum walsbyi: vanity is but the surface |journal=BMC Genomics |volume=18 |issue=1 |pages=510 |doi=10.1186/s12864-017-3892-2 |pmc=5496347 |pmid=28673248}}</ref> The second strain was an Australian isolate, called C23.<ref name=":02" /> Five strains were additionally isolated, totaling in seven total isolates of ''H. walsbyi.''<ref>{{Cite journal |last=Podell |first=Sheila |last2=Ugalde |first2=Juan A. |last3=Narasingarao |first3=Priya |last4=Banfield |first4=Jillian F. |last5=Heidelberg |first5=Karla B. |last6=Allen |first6=Eric E. |date=2013-04-18 |title=Assembly-Driven Community Genomics of a Hypersaline Microbial Ecosystem |url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3630111/ |journal=PLoS ONE |volume=8 |issue=4 |pages=e61692 |doi=10.1371/journal.pone.0061692 |issn=1932-6203 |pmc=3630111 |pmid=23637883}}</ref> In a specific hypersaline environment, Lake Tyrrell, ''Haloquadratum walsbyi'' made up nearly 38% of the community of archaea found when the ecosystem was cultured.<ref>{{Cite journal |last=Ghai |first=Rohit |last2=Pašić |first2=Lejla |last3=Fernández |first3=Ana Beatriz |last4=Martin-Cuadrado |first4=Ana-Belen |last5=Mizuno |first5=Carolina Megumi |last6=McMahon |first6=Katherine D. |last7=Papke |first7=R. Thane |last8=Stepanauskas |first8=Ramunas |last9=Rodriguez-Brito |first9=Beltran |last10=Rohwer |first10=Forest |last11=Sánchez-Porro |first11=Cristina |last12=Ventosa |first12=Antonio |last13=Rodríguez-Valera |first13=Francisco |date=2011-10-31 |title=New Abundant Microbial Groups in Aquatic Hypersaline Environments |url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3216616/ |journal=Scientific Reports |volume=1 |pages=135 |doi=10.1038/srep00135 |issn=2045-2322 |pmc=3216616 |pmid=22355652}}</ref> |
|||
In 2004, two strains of ''H. walsbyi'' were successfully isolated and able to be sequenced. The first strain is an isolate from the Spanish, called HBSQ00.<ref name=":0" /> The second strain is an Australian isolate, called C23.<ref name=":0" /> |
|||
== Normal Microbiota == |
== Normal Microbiota == |
||
Revision as of 05:16, 17 November 2022
| Haloquadratum walsbyi | |
|---|---|

| |
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | H. walsbyi
|
| Binomial name | |
| Haloquadratum walsbyi Burns et al. 2007
| |
Haloquadratum walsbyi is of the genus Haloquadratum, within the archaea domain known for its square halophilic nature.[1] First discovered in a brine pool in the Sinai peninsula of Egypt, H. walsbyi is noted for its flat, square-shaped cells, and its unusual ability to survive in aqueous environments with high concentrations of sodium chloride and magnesium chloride.[2][3] The species' genus name Haloquadratum translates from Greek and Latin as "salt square". This archaean is also commonly referred to as "Walsby's Square Bacterium" because of its identifying square shape which makes it very unique.[4] In accordance with its name, Haloquadratum walsbyi are most abundantly observed in salty environments.
Haloquadratum walsbyi is a phototrophic halophilic archaeon. It was the only recognized species of the genus Haloquadratum until 1999 when Haloarcula quadrata was reported as recovered from a brine pool.[2] Haloquadratum walsbyi is very unusual because of its unique cellular structure that resembles an almost-perfectly, flat-shaped figure.
The genus was first observed in 1980 by a British microbiologist, Prof. Anthony E. Walsby, from samples taken from the Sabkha Gavish, a brine lake in southern Sinai, Egypt. This discovery is formally described in 2007 by Burns et al. Attempts to cultivate the archaea were unsuccessful until 2004 and resulted in the identification of Haloarcula quadrata, another species of square archaea of the genus Haloarcula, distinct from H. walsbyi, less abundant and genetically quite different.[5]
Description
Haloquadratum walsbyi cells size at 2 to 5 microns and 100 to 200 nanometers thick. The archaea generally contain granules of polyhydroxyalkanoates and hold a number of refractive vacuoles filled with gas that ensure buoyancy in an aqueous environment, and allow for maximum light absorption. These gas vacuoles were discovered by Wallaby in 1980 when determining the identify of intracellular refractive bodies in the archaean's structure.[4] They gather in sheets up to 40 μm wide, but the connections between the cells are fragile and can easily be broken.[6]
These organisms can be found in any stretch of very salty water. During the evaporation of seawater, calcium carbonate (CaCO3) and calcium sulfate (CaSO4) precipitate first, leading to a brine rich in sodium chloride NaCl. If evaporation continues, NaCl precipitates in the form of halite, leaving a brine rich in magnesium chloride (MgCl2). H. walsbyi prospers during the final phase of the precipitation of halite, and can constitute 80% of the biomass of this medium.[7] Haloquadratum walsbyi cells have been determined to be Gram-negative through staining and when grown in a laboratory the best determined conditions for growth is a media with 18% salts at a neutral pH[8]
The genome of H. walsbyi has been completely sequenced, allowing access to a better understanding of the phylogenetic and taxonomic classification of this organism and its role in the ecosystem. A genomic comparison of Spanish and Australian isolates (strains HBSQ001 and C23T) strongly suggests a rapid global dispersion, as they are remarkably similar and have maintained the order of genes.[citation needed]
Its growth in the laboratory was obtained in a medium with very high chloride concentrations (greater than 2 mol · L−1 of MgCl2 and greater than 3 mol · L −1of NaCl ), making this organism among the most haloresistant known. Its optimum growth temperature is 40 °C, making this archaea a Mesophile.
-
Optical phase-contrast microscopy image of a Haloquadratum walsbyi square cell. The numerous light dots are gas vesicles that allow flotation to the surface, most likely to acquire oxygen.[9]Scale bar 1 µm
-
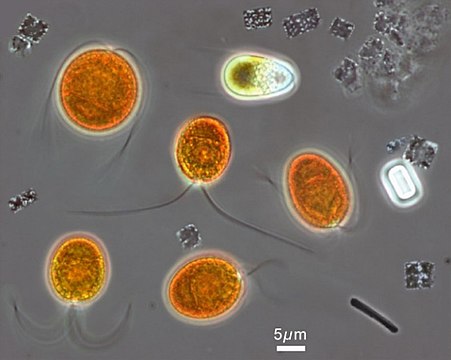
Microscopic image from the hypersaline Lake Tyrrell, in which orange chlorophyte Dunaliella salina can be tentatively identified, accompanied by a number of smaller Haloquadratum walsbyi, showing their flat square-shaped cells.
Diversity
A surprisingly high amount of cells in salt brines around the world are Haloquadratum walsbyi. Up to 80% of the cells in the brines can be identified as Haloquadratum walsbyi. Experiments have been done to examine the genetic diversity in the salt brine environment. Seven different types of H. walsbyi's genomic island have been discovered in natural environments.[10] After examining the metogenomic fosmid library for H. walsbyi, two types of the cell-wall associated islands were identified. The genes in these islands include those responsible for the synthesis of surface layer structures such as glycoproteins and genes responsible for the synthesis of cell envelopes.[4] Homologous recombination is responsible for the maintaining the genes mentioned above and also the diversity of the metagenome in its natural environment. Surface structures on different H. walsbyi cells help to differentiate sources of lineage for the population as a whole. These differing structures also increase the diversity of the cells in their natural environment. These changes in cell structure may be due to the cells' attempts to reduce their susceptibility to attack by viruses.[10]
Genomics and Structure
H. walsbyi is classified as an oligotrophic microorganism, as it grows in nutrient deficient conditions where concentrations of organic substances are minimal. To combat, H. walsbyi maintains a high surface to volume ratio by flattening to maximize nutrient uptake. Because of their square shape, they are more capable of flattening than spherical shaped microorganisms are.[3] H. walsbyi can flatten an extreme amount of about 0.1-0.5μm. The overall size of the cell structure ranges from 1.5 to 11 μm. However, larger cells have been observed. The largest recorded H. walsbyi cell was measured as 40 x 40 μm.[11]
The square shape of H. walsbyi has been the focus of many studies. It is able to maintain this structure due to its adaptive traits.[12] These traits can be found in both H. walsbyi's genome composition as well as its protein sequences. These adaptations allow H. walsbyi to thrive in environments such as saturated brines while also maintaining a defined square structure.[12]
A typical Genome of H. walsbyi has a 3,132,494 bp chromosome. The strain HBSQ001, DSM 16790 was analyzed to obtain this data. H. walsbyi is distinguished by the abnormally low Guanine-Cytosine (GC) content compared to other haloarchaea. H. walsbyi has an average of 47.9% GC content compared to the expected 60-70%. Additionally, the proteins encoded are highly conserved specifically in the amino acid sequence. It is understood that H. walsbyi evolved from a typical GC rich, moderately conserving ancestor.[3]
History
The Haloquadratum walsbyi archaea was first discovered in 1980 by a microbiology professor Anthony E. Walsby.[13] The microbe was initially named after him as “Walsby's square bacterium," as it was discovered before the archaea domain was acknowledged in full.[14] It is now formally known as Haloquadratum walsbyi, and considered a well known halophilic archaea. Additionally, it is accredited to be one of first archaea discovered with a square cellular shape.[15]
Upon the observation of the unique shape of H. walsbyi, cultivation has been a goal for scientists studying the species. Hyper-saline media has been found to be a substantial medium to maintain the pure cultures.[16] H. walsbyi remains one of the largest prokaryotes known today and contains roughly 3 million basepairs.[16]
In 2004, two strains of H. walsbyi were successfully isolated and able to be sequenced. The first strain was an isolate from the Spanish, called HBSQ00.[17] The second strain was an Australian isolate, called C23.[17] Five strains were additionally isolated, totaling in seven total isolates of H. walsbyi.[18] In a specific hypersaline environment, Lake Tyrrell, Haloquadratum walsbyi made up nearly 38% of the community of archaea found when the ecosystem was cultured.[19]
Normal Microbiota
The Archaeon Haloquadratum walsbyi is abundant in red brines, in salt lakes and solar salter crystallizer ponds,[20] shallow ponds that are connected to each other and increase in salinity.[21] Bacteriorhodopsin, a membrane protein that uses energy from light to drive the hydrogen-ion pump,[22] which are found in Haloquadratum walsbyi absorbs energy from light and are found in communities within these brines.[20] The use of these bacteriorhodopsin shows the photoheterotroph nature of Haloquadratum walsbyi. The salt saturated environments that this archaean inhabits, along with being rich in magnesium chloride, have very low activity within the water which causes desiccation stress. These salt saturated environments can have a salinity of more than ten-times that of average seawater. The magnesium saturation in these ecosystems, what is also referred to as bitterns, are most often found with very little to no life present.[23] This environment is very hostile and H. walsbyi is only able to survive in it due to its unique genomic make up and while other organisms would perish in similar conditions. [24]
References
- ^ Bolhuis, Henk; Palm, Peter; Wende, Andy; Falb, Michaela; Rampp, Markus; Rodriguez-Valera, Francisco; Pfeiffer, Friedhelm; Oesterhelt, Dieter (2006-07-04). "The genome of the square archaeon Haloquadratum walsbyi : life at the limits of water activity". BMC Genomics. 7: 169. doi:10.1186/1471-2164-7-169. ISSN 1471-2164. PMC 1544339. PMID 16820047.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Oren A, Ventosa A, Gutiérrez MC, Kamekura M (July 1999). "Haloarcula quadrata sp. nov., a square, motile archaeon isolated from a brine pool in Sinai (Egypt)". International Journal of Systematic Bacteriology. 49 (3): 1149–1155. doi:10.1099/00207713-49-3-1149. PMID 10425773.
- ^ a b c Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, et al. (July 2006). "The genome of the square archaeon Haloquadratum walsbyi : life at the limits of water activity". BMC Genomics. 7 (1): 169. doi:10.1186/1471-2164-7-169. PMC 1544339. PMID 16820047.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c "Haloquadratum walsbyi - overview". www.biochem.mpg.de. Retrieved 2022-11-16.
- ^ "Haloquadratum walsbyi articles – Encyclopedia of Life". eol.org. Retrieved 2022-11-16.
- ^ Sublimi Saponetti M, Bobba F, Salerno G, Scarfato A, Corcelli A, Cucolo A (April 2011). "Morphological and structural aspects of the extremely halophilic archaeon Haloquadratum walsbyi". PLOS ONE. 6 (4): e18653. Bibcode:2011PLoSO...618653S. doi:10.1371/journal.pone.0018653. PMC 3084702. PMID 21559517.
- ^ Cite error: The named reference
lifewas invoked but never defined (see the help page). - ^ Burns, David G.; Janssen, Peter H.; Itoh, Takashi; Kamekura, Masahiro; Li, Zhuo; Jensen, Grant; Rodríguez-Valera, Francisco; Bolhuis, Henk; Dyall-Smith, Mike L.YR 2007. "Haloquadratum walsbyi gen. nov., sp. nov., the square haloarchaeon of Walsby, isolated from saltern crystallizers in Australia and Spain". International Journal of Systematic and Evolutionary Microbiology. 57 (2): 387–392. doi:10.1099/ijs.0.64690-0. ISSN 1466-5034.
{{cite journal}}: CS1 maint: numeric names: authors list (link) - ^ Sublimi Saponetti M, Bobba F, Salerno G, Scarfato A, Corcelli A, Cucolo A (April 2011). "Morphological and structural aspects of the extremely halophilic archaeon Haloquadratum walsbyi". PLOS ONE. 6 (4): e18653. Bibcode:2011PLoSO...618653S. doi:10.1371/journal.pone.0018653. PMC 3084702. PMID 21559517.
- ^ a b Martin-Cuadrado, Ana-Belen; Pašić, Lejla; Rodriguez-Valera, Francisco (2015-08-13). "Diversity of the cell-wall associated genomic island of the archaeon Haloquadratum walsbyi". BMC Genomics. 16 (1): 603. doi:10.1186/s12864-015-1794-8. ISSN 1471-2164. PMC 4535781. PMID 26268990.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Zenke, Ralf; von Gronau, Susanne; Bolhuis, Henk; Gruska, Manuela; Pfeiffer, Friedhelm; Oesterhelt, Dieter (2015). "Fluorescence microscopy visualization of halomucin, a secreted 927 kDa protein surrounding Haloquadratum walsbyi cells". Frontiers in Microbiology. 6. doi:10.3389/fmicb.2015.00249/full. ISSN 1664-302X.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Bolhuis, Henk; Palm, Peter; Wende, Andy; Falb, Michaela; Rampp, Markus; Rodriguez-Valera, Francisco; Pfeiffer, Friedhelm; Oesterhelt, Dieter (2006-07-04). "The genome of the square archaeon Haloquadratum walsbyi : life at the limits of water activity". BMC Genomics. 7 (1): 169. doi:10.1186/1471-2164-7-169. ISSN 1471-2164. PMC 1544339. PMID 16820047.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Bolhuis H, Martín-Cuadrado AB, Rosselli R, Pašić L, Rodriguez-Valera F (July 2017). "Transcriptome analysis of Haloquadratum walsbyi: vanity is but the surface". BMC Genomics. 18 (1): 510. doi:10.1186/s12864-017-3892-2. PMC 5496347. PMID 28673248.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Legault BA, Lopez-Lopez A, Alba-Casado JC, Doolittle WF, Bolhuis H, Rodriguez-Valera F, Papke RT (July 2006). "Environmental genomics of "Haloquadratum walsbyi" in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species". BMC Genomics. 7 (1): 171. doi:10.1186/1471-2164-7-171. PMC 1560387. PMID 16820057.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lobasso S, Lopalco P, Mascolo G, Corcelli A (December 2008). "Lipids of the ultra-thin square halophilic archaeon Haloquadratum walsbyi". Archaea. 2 (3): 177–183. doi:10.1155/2008/870191. PMC 2685597. PMID 19054744.
- ^ a b Bolhuis, Henk; Poele, Evelien M. te; Rodriguez-Valera, Francisco (24 September 2004). "Isolation and cultivation of Walsby's square archaeon". Environmental Microbiology. 6 (12): 1287–1291. doi:10.1111/j.1462-2920.2004.00692.x. ISSN 1462-2912.
- ^ a b Bolhuis H, Martín-Cuadrado AB, Rosselli R, Pašić L, Rodriguez-Valera F (July 2017). "Transcriptome analysis of Haloquadratum walsbyi: vanity is but the surface". BMC Genomics. 18 (1): 510. doi:10.1186/s12864-017-3892-2. PMC 5496347. PMID 28673248.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Podell, Sheila; Ugalde, Juan A.; Narasingarao, Priya; Banfield, Jillian F.; Heidelberg, Karla B.; Allen, Eric E. (2013-04-18). "Assembly-Driven Community Genomics of a Hypersaline Microbial Ecosystem". PLoS ONE. 8 (4): e61692. doi:10.1371/journal.pone.0061692. ISSN 1932-6203. PMC 3630111. PMID 23637883.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Ghai, Rohit; Pašić, Lejla; Fernández, Ana Beatriz; Martin-Cuadrado, Ana-Belen; Mizuno, Carolina Megumi; McMahon, Katherine D.; Papke, R. Thane; Stepanauskas, Ramunas; Rodriguez-Brito, Beltran; Rohwer, Forest; Sánchez-Porro, Cristina; Ventosa, Antonio; Rodríguez-Valera, Francisco (2011-10-31). "New Abundant Microbial Groups in Aquatic Hypersaline Environments". Scientific Reports. 1: 135. doi:10.1038/srep00135. ISSN 2045-2322. PMC 3216616. PMID 22355652.
- ^ a b "Haloquadratum walsbyi - an overview | ScienceDirect Topics". www.sciencedirect.com. Retrieved 2022-10-27.
- ^ Antón, Josefa; Rosselló-Mora, Ramón; Rodríguez-Valera, Francisco; Amann, Rudolf (July 2000). "Extremely Halophilic Bacteria in Crystallizer Ponds from Solar Salterns". Applied and Environmental Microbiology. 66 (7): 3052–3057. Bibcode:2000ApEnM..66.3052A. doi:10.1128/aem.66.7.3052-3057.2000. ISSN 0099-2240. PMC 92110. PMID 10877805.
- ^ Henderson, R.; Schertler, G. F. (1990-01-30). "The structure of bacteriorhodopsin and its relevance to the visual opsins and other seven-helix G-protein coupled receptors". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 326 (1236): 379–389. Bibcode:1990RSPTB.326..379H. doi:10.1098/rstb.1990.0019. ISSN 0962-8436. PMID 1970644.
- ^ Oren, A (2002-01). "Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications". Journal of Industrial Microbiology & Biotechnology. 28 (1): 56–63. doi:10.1038/sj/jim/7000176.
{{cite journal}}: Check date values in:|date=(help) - ^ Bolhuis, Henk; Palm, Peter; Wende, Andy; Falb, Michaela; Rampp, Markus; Rodriguez-Valera, Francisco; Pfeiffer, Friedhelm; Oesterhelt, Dieter (2006-07-04). "The genome of the square archaeon Haloquadratum walsbyi : life at the limits of water activity". BMC Genomics. 7: 169. doi:10.1186/1471-2164-7-169. ISSN 1471-2164. PMC 1544339. PMID 16820047.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
Further reading
- Lobasso S, Lopalco P, Vitale R, Saponetti MS, Capitanio G, Mangini V, et al. (9 February 2012). "The light-activated proton pump Bop I of the archaeon Haloquadratum walsbyi". Photochemistry and Photobiology. 88 (3): 690–700. doi:10.1111/j.1751-1097.2012.01089.x. PMID 22248212. S2CID 41934280.
- Cuebas-Irizarry MF, Irizarry-Caro RA, López-Morales C, Badillo-Rivera KM, Rodríguez-Minguela CM, Montalvo-Rodríguez R (November 2017). "Cloning and Molecular Characterization of an Alpha-Glucosidase (MalH) from the Halophilic Archaeon Haloquadratum walsbyi". Life. 7 (4): 46. doi:10.3390/life7040046. PMC 5745559. PMID 29160840.

![Optical phase-contrast microscopy image of a Haloquadratum walsbyi square cell. The numerous light dots are gas vesicles that allow flotation to the surface, most likely to acquire oxygen.[9] Scale bar 1 µm](http://upload.wikimedia.org/wikipedia/commons/thumb/1/14/Optical_phase-contrast_microscopy_image_of_a_Haloquadratum_walsbyi_square_cell_-_PLoS_ONE.png/374px-Optical_phase-contrast_microscopy_image_of_a_Haloquadratum_walsbyi_square_cell_-_PLoS_ONE.png)