Frameshift mutation: Difference between revisions
DougCovert (talk | contribs) minor title change |
DougCovert (talk | contribs) Added Diseases SMS, Sudden death, Huntington |
||
| Line 31: | Line 31: | ||
==Frameshift Mutation Causes-Research== |
==Frameshift Mutation Causes-Research== |
||
====Finding Frameshift Mutations==== |
====Finding Frameshift Mutations==== |
||
=====Fluorescence===== |
|||
The effects of neighboring bases and secondary structure on the frequency of frameshift mutations has been investigated in depth. Fluorescently tagged DNA, by means of base analogues, permits one to study the local changes of a DNA sequence. <ref>{{cite |first=Neil P. |last=Johnson |coauthors=Walter A. Baase, Peter H. von Hippel |title=Low-energy circular dichroism of 2-aminopurine dinucleotide as a probe of local conformation of DNA and RNA |quote=PNAS 2004 101:3426-3431; published online before print March 1, 2004 |doi=10.1073/pnas.0400591101}}</ref> Studies on the effects of the length of the primer strand reveal that an equilibrium mixture of four hybridization conformations was observed when template bases looped-out as a bulge, i.e. a structure flanked on both sides by duplex DNA. In contrast, a double-loop structure with an unusual unstacked DNA conformation at its downstream edge was observed when the extruded bases were positioned at the primer–template junction, showing that misalignments can be modified by neighboring DNA secondary structure.<ref>{{cite |first=Walter A. |last=Baase |coauthors=Davis Jose , Benjamin C. Ponedel , Peter H. von Hippel , and Neil P. Johnson |title=DNA models of trinucleotide frameshift deletions: the formation of loops and bulges at the primer–template junction |quote=Nucleic Acids Research Advance Access published on April 1, 2009 |doi=10.1093/nar/gkn1042 |periodical=Nucleic Acids Research |volume=37 |issue=5 |pages=1682–1689}} </ref> |
The effects of neighboring bases and secondary structure on the frequency of frameshift mutations has been investigated in depth. Fluorescently tagged DNA, by means of base analogues, permits one to study the local changes of a DNA sequence. <ref>{{cite |first=Neil P. |last=Johnson |coauthors=Walter A. Baase, Peter H. von Hippel |title=Low-energy circular dichroism of 2-aminopurine dinucleotide as a probe of local conformation of DNA and RNA |quote=PNAS 2004 101:3426-3431; published online before print March 1, 2004 |doi=10.1073/pnas.0400591101}}</ref> Studies on the effects of the length of the primer strand reveal that an equilibrium mixture of four hybridization conformations was observed when template bases looped-out as a bulge, i.e. a structure flanked on both sides by duplex DNA. In contrast, a double-loop structure with an unusual unstacked DNA conformation at its downstream edge was observed when the extruded bases were positioned at the primer–template junction, showing that misalignments can be modified by neighboring DNA secondary structure.<ref>{{cite |first=Walter A. |last=Baase |coauthors=Davis Jose , Benjamin C. Ponedel , Peter H. von Hippel , and Neil P. Johnson |title=DNA models of trinucleotide frameshift deletions: the formation of loops and bulges at the primer–template junction |quote=Nucleic Acids Research Advance Access published on April 1, 2009 |doi=10.1093/nar/gkn1042 |periodical=Nucleic Acids Research |volume=37 |issue=5 |pages=1682–1689}} </ref> |
||
Sanger sequencing and pyrosequencing are two methods that have been used to detect frameshift mutations, however, it is likely that data generated will not be of the highest quality. Even still, 1.96 million indels have been identified through Sanger sequencing that do not overlap with other databases. When a frameshift mutation is observed it is compared against the Human Genome Mutation Database (HGMD) to determine if the mutation has a damaging effect. This is done by looking at four features. First, the ratio between the affected and conserved DNA, second the location of the mutation relative to the transcript, third the ratio of conserved and affected amino acids and finally the distance of the indel to the end of the exon. <ref name="predicting frameshifts" /> |
Sanger sequencing and pyrosequencing are two methods that have been used to detect frameshift mutations, however, it is likely that data generated will not be of the highest quality. Even still, 1.96 million indels have been identified through Sanger sequencing that do not overlap with other databases. When a frameshift mutation is observed it is compared against the Human Genome Mutation Database (HGMD) to determine if the mutation has a damaging effect. This is done by looking at four features. First, the ratio between the affected and conserved DNA, second the location of the mutation relative to the transcript, third the ratio of conserved and affected amino acids and finally the distance of the indel to the end of the exon. <ref name="predicting frameshifts" /> |
||
=====Ribosomal Slippage===== |
|||
[[Huntington’s disease]] is one of the nine codon reiteration disorders caused by polyglutamine expansion mutations that include spino-cerebellar ataxia (SCA) 1, 2, 6, 7 and 3, spinobulbar muscular atrophy and dentatorubal-pallidoluysianatrophy. There may be a link between diseases caused by polyglutamine and polyalanine expansion mutations, as frame shifting of the original SCA3 gene product encoding CAG/polyglutamines to GCA/polyalanines. Ribosomal slippage during translation of the SCA3 protein has been proposed as the mechanism resulting in shifting from the polyglutamine to the polyalanine-encoding frame. A dinucleotide deletion or single nucleotide insertion within the polyglutamine tract of huntingtin exon 1 would shift the CAG, polyglutamineen coding frame by +1 (+1 frame shift) to the GCA, polyalanine-encoding frame and introduce a novel epitope to the C terminus of Htt exon 1 (APAAAPAATRPGCG).<ref>{{cite journal|last=Davies|first=J E|coauthors=Rubinsztein, D C|title=Polyalanine and polyserine frameshift products in Huntington's disease|journal=Journal of Medical Genetics|date=NaN undefined NaN|volume=43|issue=11|pages=893–896|doi=10.1136/jmg.2006.044222}}</ref> |
|||
====Research in Frequency of Frameshift Mutations==== |
====Research in Frequency of Frameshift Mutations==== |
||
Frameshift mutations are found to be more common in repeat regions of DNA. A reason for this is because of slipping of the polymerase enzyme in repeat regions, allowing for mutations to enter the sequence. <ref name="editing frameshift mutation">{{cite journal|last=Harfe|first=BD|coauthors=Jinks-Robertson, S|title=Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae.|journal=Molecular and cellular biology|date=1999 Jul|volume=19|issue=7|pages=4766-73|pmid=10373526|accessdate=20 March 2013}}</ref> Experiments can be run to determine the frequency of the frameshift mutation by adding or removing a pre-set number of nucleotides. Experiements have been run by adding four basepairs, called the +4 experiments, but a team from Emory University looked at the difference in frequency of the mutation by both adding and deleting a base pair. It was shown that there was no difference in the frequency between the addition and deletion of a base pair. There is however, a difference in the end result of the protein. <ref name="editing frameshift mutation" /> |
Frameshift mutations are found to be more common in repeat regions of DNA. A reason for this is because of slipping of the polymerase enzyme in repeat regions, allowing for mutations to enter the sequence. <ref name="editing frameshift mutation">{{cite journal|last=Harfe|first=BD|coauthors=Jinks-Robertson, S|title=Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae.|journal=Molecular and cellular biology|date=1999 Jul|volume=19|issue=7|pages=4766-73|pmid=10373526|accessdate=20 March 2013}}</ref> Experiments can be run to determine the frequency of the frameshift mutation by adding or removing a pre-set number of nucleotides. Experiements have been run by adding four basepairs, called the +4 experiments, but a team from Emory University looked at the difference in frequency of the mutation by both adding and deleting a base pair. It was shown that there was no difference in the frequency between the addition and deletion of a base pair. There is however, a difference in the end result of the protein. <ref name="editing frameshift mutation" /> |
||
| Line 48: | Line 50: | ||
====Cystic Fibrosis==== |
====Cystic Fibrosis==== |
||
[[Cystic Fibrosis]] (CF) is a disease based on mutations in the CF transmembrane conductance regulator (CFTR) gene. There are over 1500 mutations identified, but not all cause the disease. <ref name="guidelines for CF">{{cite journal|last=Farrell|first=Philip M.|coauthors=Rosenstein, Beryl J.; White, Terry B.; Accurso, Frank J.; Castellani, Carlo; Cutting, Garry R.; Durie, Peter R.; LeGrys, Vicky A.; Massie, John; Parad, Richard B.; Rock, Michael J.; Campbell, Preston W.|title=Guidelines for Diagnosis of Cystic Fibrosis in Newborns through Older Adults: Cystic Fibrosis Foundation Consensus Report|journal=The Journal of Pediatrics|date=NaN undefined NaN|volume=153|issue=2|pages=S4–S14|doi=10.1016/j.jpeds.2008.05.005|accessdate=21 March 2013}}</ref> Most cases of cystic fibrosis are a result of the ∆F508 mutation, which deletes the entire amino acid. Two frameshift mutations are of interest in diagnosing CF, CF1213delT and CF1154-insTC. Both of these mutations commonly occur in tandem with at least one other mutation. They both lead to a small decrease in the function of the lungs and occur in about 1% of patients tested. These mutations were identified through Sanger sequencing. <ref name="frameshift mutations in CF">{{cite journal|last=Iannuzzi|first=MC|coauthors=Stern, RC; Collins, FS; Hon, CT; Hidaka, N; Strong, T; Becker, L; Drumm, ML; White, MB; Gerrard, B|title=Two frameshift mutations in the cystic fibrosis gene.|journal=American journal of human genetics|date=1991 Feb|volume=48|issue=2|pages=227-31|pmid=1990834|accessdate=21 March 2013}}</ref> |
[[Cystic Fibrosis]] (CF) is a disease based on mutations in the CF transmembrane conductance regulator (CFTR) gene. There are over 1500 mutations identified, but not all cause the disease. <ref name="guidelines for CF">{{cite journal|last=Farrell|first=Philip M.|coauthors=Rosenstein, Beryl J.; White, Terry B.; Accurso, Frank J.; Castellani, Carlo; Cutting, Garry R.; Durie, Peter R.; LeGrys, Vicky A.; Massie, John; Parad, Richard B.; Rock, Michael J.; Campbell, Preston W.|title=Guidelines for Diagnosis of Cystic Fibrosis in Newborns through Older Adults: Cystic Fibrosis Foundation Consensus Report|journal=The Journal of Pediatrics|date=NaN undefined NaN|volume=153|issue=2|pages=S4–S14|doi=10.1016/j.jpeds.2008.05.005|accessdate=21 March 2013}}</ref> Most cases of cystic fibrosis are a result of the ∆F508 mutation, which deletes the entire amino acid. Two frameshift mutations are of interest in diagnosing CF, CF1213delT and CF1154-insTC. Both of these mutations commonly occur in tandem with at least one other mutation. They both lead to a small decrease in the function of the lungs and occur in about 1% of patients tested. These mutations were identified through Sanger sequencing. <ref name="frameshift mutations in CF">{{cite journal|last=Iannuzzi|first=MC|coauthors=Stern, RC; Collins, FS; Hon, CT; Hidaka, N; Strong, T; Becker, L; Drumm, ML; White, MB; Gerrard, B|title=Two frameshift mutations in the cystic fibrosis gene.|journal=American journal of human genetics|date=1991 Feb|volume=48|issue=2|pages=227-31|pmid=1990834|accessdate=21 March 2013}}</ref> |
||
====Tay-Sachs Disease==== |
|||
A severe genetic disease with no known cure..... |
|||
====Smith-Magenis Syndrome==== |
|||
Smith-Magenis syndrome (SMS)is a complex syndrome involving intellectual disabilities, sleep disturbance, behavioural problems, and a variety of craniofacial, skeletal, and visceral anomalies. The majority of SMS cases harbor an ~3.5 Mb common deletion that encompasses the retinoic acid induced-1 (RAI1) gene. Other cases illustrate variability in the SMS phenotype not previously shown for RAI1 mutation,including hearing loss, absence of self-abusive behaviours, and mild global delays.Sequencing of RAI1 revealed mutation of a heptamericC-tract (CCCCCCC) in exon 3 resulting in frameshift mutations. Of the seven reported frameshift mutations occurring in poly C-tracts in RAI1, four cases (~57%) occur at this heptameric C-tract. The results indicate that this heptameric C-tract is a preferential hotspot insertion/deletions (SNindels) and therefore a primary target for analysis in patients suspected for mutations in RAI1.<ref>{{cite journal|last=Truong|first=Hoa T|coauthors=Dudding, Tracy; Blanchard, Christopher L.; Elsea, Sarah H|title=Frameshift mutation hotspot identified in Smith-Magenis syndrome: case report and review of literature|journal=BMC Medical Genetics|date=NaN undefined NaN|volume=11|issue=1|pages=142|doi=10.1186/1471-2350-11-142}}</ref> |
|||
====Hypertrophic Cardiomyopathy==== |
|||
Hypertrophic cardiomyopathy is the most common cause of sudden death in young people, including trained athletes, and is caused by mutations in genes encoding proteins of the cardiac sarcomere. Mutations in the Troponin C gene (TNNC1) are a rare genetic cause of hypertrophic cardiomyopathy. A recent study has indicated that a frameshift mutation (c.363dupG or p.Gln122AlafsX30) in Troponin C was the cause of hypertrophic cardiomyopathy (and sudden cardiac death) in a 19-year-old male.<ref name="pmid21262074">{{cite journal |author=Chung WK, Kitner C, Maron BJ |title=Novel frameshift mutation in Troponin C ( TNNC1) associated with hypertrophic cardiomyopathy and sudden death |journal=Cardiol Young |volume=21 |issue=3 |pages=345–8 |year=2011 |month=June |pmid=21262074 |doi=10.1017/S1047951110001927 |url=}}</ref> |
|||
===Cures=== |
===Cures=== |
||
Revision as of 15:19, 22 March 2013
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels (insertions or deletions) of a number of nucleotides that is not evenly divisible by three from a DNA sequence. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame (the grouping of the codons), resulting in a completely different translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein.[1]

A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon ("UAA", "UGA" or "UAG") encountered in the sequence. The polypeptide being created could be abnormally short or abnormally long, and will most likely not be functional.
Frameshift mutations frequently result in severe genetic diseases such as Tay-Sachs disease. A frameshift mutation is responsible for the disabling of the CCR5 HIV receptor and some types of familial hypercholesterolemia (Lewis, 2005, p. 227-228). Frameshift mutations have also been proposed as a source of biological novelty, as with the alleged creation of nylonase. However, a study by Negoro et al (2006) [2] found that a frameshift mutation was unlikely to have been the cause and that rather a two amino acid substitution in the catalytic cleft of an ancestral esterase amplified Ald-hydrolytic activity.
Frameshifting may also occur during prophase translation, producing different proteins from overlapping open reading frames, such as the gag-pol-env retroviral proteins. This is fairly common in viruses and also occurs in bacteria and yeast (Farabaugh, 1996). Reverse transcriptase, as opposed to RNA Polymerase II, is thought to be a stronger cause of the occurrence of frameshift mutations. In experiments only 3-13% of all frameshift mutations occured because of RNA Polymerase II. In prokaryotes the error rate inducing frameshift mutations is only somewhere in the range of .0001 and .00001. [3]
A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted.
Molecular Biology and Biochemistry Background
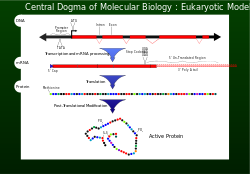
Central Dogma
In 1956 Francis Crick described the flow of genetic information from DNA to a specific amino acid arrangement for making a protein as the central dogma.[1] For a cell to properly function, proteins are required to be produced accurately for structural and for catalytic activities. An incorrectly made protein can have determinental effects on cell viability and in most cases cause the higher organism to become unhealthy by abnormal cellular functions. To ensure that the genome successfully passes the information on, proofreading mechanisms such as exonucleases and mismatch repair systems are incorporated in DNA replication .[1]
Transcription and Translation

After DNA replication, the reading of a selected section of genetic information is accomplished by transcription.[1] Nucleotides containing the genetic information is now on a single strand messenger template called mRNA. The mRNA is incorporated with a subunit of the ribosome and interacts with an rRNA. This association along with aminoacyl-tRNA binding to the start codon (AUG) initiates translation. The genetic information carried in the codons of the mRNA are now read (decoded) by anticodons of the tRNA. As each codon (triplet) is read, amino acids are being joined together until a stop codon (UAG, UGA,or UAA) is reached. At this point the polypeptide (protein) has been synthesised and is released. [1] For every 1000 amino acid incorporated into the protein, no more than one is incorrect. This fidelity of codon recognition is maintained by proper base pairing at the ribosome A site, GTP hydrolysis activity of EF-Tu a form of kinetic stability, and a proofreading mechanism as EF-Tu is released. [1]
Codon-triplet importance

A codon is a set of three nucleotides, a triplet that code for a certain amino acid. The first codon establishes the reading frame, whereby a new codon begins. A proteins amino acid backbone sequence is defined by contiguous triplets.[4]Codons are key to translation of genetic information for the synthesis of proteins. The reading frame is set when translating themRNA begins and is maintained as it reads one triplet to the next. The reading of the genetic code is subject to three rules the monitor codons in mRNA. First, condons are read in a 5' to 3' direction. Second, codons are nonoverlapping and the message has no gaps. The last rule, as stated above, that the message is translated in a fixed reading frame. [1]
Mechanism
The Mutation
The cell has various ways to ensure the correct transfer of genetic information beginning at DNA replication, during translation and rules that govern the genetic code. How does a mutation alter the genetic code. Frameshift mutation is not the only type. There are two other types of point mutations, namely a missense mutation and a nonsense mutation. A missense mutation is where a codon specific for one amino acid is changed to a codon specific for another amino acid. Sickle-cell anemia is a classic genetic dsieases caused by this. The nonsense mutation is a change to the chain-termination codon, causing a incomplete polypeptide to be released and rapidly degraded by mRNA decay. [1] Frameshift mutations are insertions or deletions one or a small number of bases that alter the reading frame. It drastically changes the coding capacity of the message(genetic information).[1] Small insertions or deletions (those less than 20 base pairs) make up 24% of mutations in disease. [5]
Genetic or Environmental
Obviously this is a genetic mutation at the level of nucleotide bases. Why and how frameshift mutations occur are continually being sought after.
Frameshift Mutation Causes-Research
Finding Frameshift Mutations
Fluorescence
The effects of neighboring bases and secondary structure on the frequency of frameshift mutations has been investigated in depth. Fluorescently tagged DNA, by means of base analogues, permits one to study the local changes of a DNA sequence. [6] Studies on the effects of the length of the primer strand reveal that an equilibrium mixture of four hybridization conformations was observed when template bases looped-out as a bulge, i.e. a structure flanked on both sides by duplex DNA. In contrast, a double-loop structure with an unusual unstacked DNA conformation at its downstream edge was observed when the extruded bases were positioned at the primer–template junction, showing that misalignments can be modified by neighboring DNA secondary structure.[7] Sanger sequencing and pyrosequencing are two methods that have been used to detect frameshift mutations, however, it is likely that data generated will not be of the highest quality. Even still, 1.96 million indels have been identified through Sanger sequencing that do not overlap with other databases. When a frameshift mutation is observed it is compared against the Human Genome Mutation Database (HGMD) to determine if the mutation has a damaging effect. This is done by looking at four features. First, the ratio between the affected and conserved DNA, second the location of the mutation relative to the transcript, third the ratio of conserved and affected amino acids and finally the distance of the indel to the end of the exon. [5]
Ribosomal Slippage
Huntington’s disease is one of the nine codon reiteration disorders caused by polyglutamine expansion mutations that include spino-cerebellar ataxia (SCA) 1, 2, 6, 7 and 3, spinobulbar muscular atrophy and dentatorubal-pallidoluysianatrophy. There may be a link between diseases caused by polyglutamine and polyalanine expansion mutations, as frame shifting of the original SCA3 gene product encoding CAG/polyglutamines to GCA/polyalanines. Ribosomal slippage during translation of the SCA3 protein has been proposed as the mechanism resulting in shifting from the polyglutamine to the polyalanine-encoding frame. A dinucleotide deletion or single nucleotide insertion within the polyglutamine tract of huntingtin exon 1 would shift the CAG, polyglutamineen coding frame by +1 (+1 frame shift) to the GCA, polyalanine-encoding frame and introduce a novel epitope to the C terminus of Htt exon 1 (APAAAPAATRPGCG).[8]
Research in Frequency of Frameshift Mutations
Frameshift mutations are found to be more common in repeat regions of DNA. A reason for this is because of slipping of the polymerase enzyme in repeat regions, allowing for mutations to enter the sequence. [9] Experiments can be run to determine the frequency of the frameshift mutation by adding or removing a pre-set number of nucleotides. Experiements have been run by adding four basepairs, called the +4 experiments, but a team from Emory University looked at the difference in frequency of the mutation by both adding and deleting a base pair. It was shown that there was no difference in the frequency between the addition and deletion of a base pair. There is however, a difference in the end result of the protein. [9]
Diseases-Research


Types
Cancer
Frameshift mutations are known to be a factor in colorectal cancer as well as other cancers with microsatellite instability. As stated previously, frameshift mutations are more likely to occur in a region of repeat sequence. When DNA mismatch repair does not fix the addition or deletion of bases, these mutations are more likely to be pathogenic. This may be in part because the tumor is not told to stop growing. Experiments in yeast and bacteria help to show characteristics of microsatellites that may contribute to defective DNA mismatch repair. These include the length of the microsatellite, the makeup of the genetic material and how pure the repeats are. Based on experimental results longer microsatellites have a higher rate of frameshift mutations. The flanking DNA can also contribute to frameshift mutations. [10] In prostate cancer a frameshift mutation changes the open reading frame (ORF) and prevents apoptosis from occuring. This leads to an unregulated growth of the tumor. While there are environmental factors that contribute to the progression of prostate cancer, there is also a genetic component. During testing of coding regions to identify mutations, 116 genetic varients were discovered, including 61 frameshift mutations. [11]
Crohn's Disease
Crohn's Disease has an association with the NOD2 gene. A frameshift mutation within the coding region of the gene can be a factor in Crohn's Disease. The mutation is an isertion of a Cytosine at position 3020. This leads to a premature stop codon, shortening the protein that is supposed to be transcribed. When the protein is able to form normally, it responds to bacterial liposaccharides, where the 3020insC mutation prevents the protein from being responsive. [12]
Cystic Fibrosis
Cystic Fibrosis (CF) is a disease based on mutations in the CF transmembrane conductance regulator (CFTR) gene. There are over 1500 mutations identified, but not all cause the disease. [13] Most cases of cystic fibrosis are a result of the ∆F508 mutation, which deletes the entire amino acid. Two frameshift mutations are of interest in diagnosing CF, CF1213delT and CF1154-insTC. Both of these mutations commonly occur in tandem with at least one other mutation. They both lead to a small decrease in the function of the lungs and occur in about 1% of patients tested. These mutations were identified through Sanger sequencing. [14]
Tay-Sachs Disease
A severe genetic disease with no known cure.....
Smith-Magenis Syndrome
Smith-Magenis syndrome (SMS)is a complex syndrome involving intellectual disabilities, sleep disturbance, behavioural problems, and a variety of craniofacial, skeletal, and visceral anomalies. The majority of SMS cases harbor an ~3.5 Mb common deletion that encompasses the retinoic acid induced-1 (RAI1) gene. Other cases illustrate variability in the SMS phenotype not previously shown for RAI1 mutation,including hearing loss, absence of self-abusive behaviours, and mild global delays.Sequencing of RAI1 revealed mutation of a heptamericC-tract (CCCCCCC) in exon 3 resulting in frameshift mutations. Of the seven reported frameshift mutations occurring in poly C-tracts in RAI1, four cases (~57%) occur at this heptameric C-tract. The results indicate that this heptameric C-tract is a preferential hotspot insertion/deletions (SNindels) and therefore a primary target for analysis in patients suspected for mutations in RAI1.[15]
Hypertrophic Cardiomyopathy
Hypertrophic cardiomyopathy is the most common cause of sudden death in young people, including trained athletes, and is caused by mutations in genes encoding proteins of the cardiac sarcomere. Mutations in the Troponin C gene (TNNC1) are a rare genetic cause of hypertrophic cardiomyopathy. A recent study has indicated that a frameshift mutation (c.363dupG or p.Gln122AlafsX30) in Troponin C was the cause of hypertrophic cardiomyopathy (and sudden cardiac death) in a 19-year-old male.[16]
Cures
A primary immunodeficiency (PID) is an inherited condition which can lead to an increase in infections. There are 120 genes and 150 mutations that play a role in primary immunodeficiencies. The standard treatment is currently gene therapy, but this is a highly risky treatment and can often lead to other diseases, such as leukemia. Gene therapy proceures include modifying the zinc fringer nuclease fustion protein, cleaving both ends of the mutation, which in turn removes it from the sequence. Antisense-oligonucleotide mediated exon skipping is another possibility for Duchnne Muscular Dystrophy. This process allows for passing over the mutation so that the rest of the sequence remains in frame and the function of the protein stays in tact. This, however, does not cure the disease, just treats symptoms and is only practical in structural proteins or other repetitive genes. A third form of repair is revertant mosaicism, which is naturally occuring by creating a reverse mutation or a mutation at a second site that corrects the reading frame. This reversion may happen by intragenic recombination, mitotic gene conversion, second site DNA slipping or site-specific reversion. This is possible in several diseases, such as X-linked Severe Combined Immunodeficiency (SCID), Wiskott-Aldrich syndrome, and Bloom syndrome. There are no drugs or other pharmacogenomic methods that help with PIDs. [17]
See also
- Translational frameshift
- Mutation
- Transcription (genetics)
- Translation (biology)
- codon
- protein
- reading frame
- point mutation
- Crohn's Disease
- Tay-Sachs Disease
References
- ^ a b c d e f g h i Losick, Richard; Watson, James D.; Tania A. Baker; Bell, Stephen; Gann, Alexander; Levine, Michael W. (2008). Molecular biology of the gene. San Francisco: Pearson/Benjamin Cummings. ISBN 0-8053-9592-X.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ http://www.jbc.org/content/280/47/39644.full.pdf+html
- ^ Zhang, J (2004 Aug). "Host RNA polymerase II makes minimal contributions to retroviral frame-shift mutations". The Journal of general virology. 85 (Pt 8): 2389–95. PMID 15269381.
{{cite journal}}:|access-date=requires|url=(help); Check date values in:|date=(help) - ^ Cox, Michael; Nelson, David R.; Lehninger, Albert L (2008). Lehninger principles of biochemistry. San Francisco: W.H. Freeman. ISBN 0-7167-7108-X.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b Hu, J (2012 Feb 9). "Predicting the effects of frameshifting indels". Genome biology. 13 (2): R9. PMID 22322200.
{{cite journal}}:|access-date=requires|url=(help); Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Johnson, Neil P., Low-energy circular dichroism of 2-aminopurine dinucleotide as a probe of local conformation of DNA and RNA, doi:10.1073/pnas.0400591101,
PNAS 2004 101:3426-3431; published online before print March 1, 2004
{{citation}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Baase, Walter A., "DNA models of trinucleotide frameshift deletions: the formation of loops and bulges at the primer–template junction", Nucleic Acids Research, vol. 37, no. 5, pp. 1682–1689, doi:10.1093/nar/gkn1042,
Nucleic Acids Research Advance Access published on April 1, 2009
{{citation}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Davies, J E (NaN undefined NaN). "Polyalanine and polyserine frameshift products in Huntington's disease". Journal of Medical Genetics. 43 (11): 893–896. doi:10.1136/jmg.2006.044222.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Harfe, BD (1999 Jul). "Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae". Molecular and cellular biology. 19 (7): 4766–73. PMID 10373526.
{{cite journal}}:|access-date=requires|url=(help); Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Schmoldt, A (1975 Sep 1). "Digitoxin metabolism by rat liver microsomes". Biochemical pharmacology. 24 (17): 1639–41. PMID 10.1093/hmg/ddq151.
{{cite journal}}:|access-date=requires|url=(help); Check|pmid=value (help); Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Xu, XiaoLin (NaN undefined NaN). "Identification of somatic mutations in human prostate cancer by RNA-Seq". Gene. doi:10.1016/j.gene.2013.01.046.
{{cite journal}}:|access-date=requires|url=(help); Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ . PMID 11385577.
{{cite journal}}:|access-date=requires|url=(help); Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ Farrell, Philip M. (NaN undefined NaN). "Guidelines for Diagnosis of Cystic Fibrosis in Newborns through Older Adults: Cystic Fibrosis Foundation Consensus Report". The Journal of Pediatrics. 153 (2): S4–S14. doi:10.1016/j.jpeds.2008.05.005.
{{cite journal}}:|access-date=requires|url=(help); Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Iannuzzi, MC (1991 Feb). "Two frameshift mutations in the cystic fibrosis gene". American journal of human genetics. 48 (2): 227–31. PMID 1990834.
{{cite journal}}:|access-date=requires|url=(help); Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Truong, Hoa T (NaN undefined NaN). "Frameshift mutation hotspot identified in Smith-Magenis syndrome: case report and review of literature". BMC Medical Genetics. 11 (1): 142. doi:10.1186/1471-2350-11-142.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help)CS1 maint: unflagged free DOI (link) - ^ Chung WK, Kitner C, Maron BJ (2011). "Novel frameshift mutation in Troponin C ( TNNC1) associated with hypertrophic cardiomyopathy and sudden death". Cardiol Young. 21 (3): 345–8. doi:10.1017/S1047951110001927. PMID 21262074.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hu, Hailiang (NaN undefined NaN). "New approaches to treatment of primary immunodeficiencies: fixing mutations with chemicals". Current Opinion in Allergy and Clinical Immunology. 8 (6): 540–546. doi:10.1097/ACI.0b013e328314b63b.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help)
Further reading
- Farabaugh PJ (1996). "Programmed translational frameshifting". Annu. Rev. Genet. 30 (1): 507–28. doi:10.1146/annurev.genet.30.1.507. PMID 8982463.
- Lewis, Ricki (2005). Human Genetics: Concepts and Applications (6th ed.). Boston, Mass: McGraw Hill. pp. 227–228. ISBN 0-07-111156-5.
- Nylonase Enzymes, 04-2004, retrieved 02-06-2009
{{citation}}: Check date values in:|accessdate=and|date=(help)
External links
- Frameshift+Mutation at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- NCBI dbSNP database — "a central repository for both single base nucleotide substitutions and short deletion and insertion polymorphisms"
- Wise2 - aligns a protein against a DNA sequence allowing frameshifts and introns
- FastY - compare a DNA sequence to a protein sequence database, allowing gaps and frameshifts
- Path - tool that compares two frameshift proteins (back-translation principle)
