Neutral network (evolution): Difference between revisions
some more references. |
|||
| Line 1: | Line 1: | ||
{{distinguish|Network-neutral data center}} |
{{distinguish|Network-neutral data center}} |
||
{{refimprove|date=January 2014}} |
|||
A '''Neutral network''' is a set of [[gene]]s all related by [[point mutation]]s that have equivalent function or [[fitness (biology)|fitness]].<ref>{{cite journal|last=van Nimwegen|first=E|coauthors=Crutchfield, JP; Huynen, M|title=Neutral evolution of mutational robustness.|journal=Proceedings of the National Academy of Sciences of the United States of America|date=1999 Aug 17|volume=96|issue=17|pages=9716–20|pmid=10449760}}</ref> Each node represents a gene sequence and each line represents the mutation connecting two sequences. Neutral networks an be though of as high, flat plateaus in a [[fitness landscape]]. During [[Neutral theory of molecular evolution|neutral evolution]], genes can randomly move through neutral networks and traverse regions of [[sequence space (evolution)|sequence space]] which may have consequences for [[robustness (evolution)|robustness]] and [[evolvability]]. |
A '''Neutral network''' is a set of [[gene]]s all related by [[point mutation]]s that have equivalent function or [[fitness (biology)|fitness]].<ref>{{cite journal|last=van Nimwegen|first=E|coauthors=Crutchfield, JP; Huynen, M|title=Neutral evolution of mutational robustness.|journal=Proceedings of the National Academy of Sciences of the United States of America|date=1999 Aug 17|volume=96|issue=17|pages=9716–20|pmid=10449760}}</ref> Each node represents a gene sequence and each line represents the mutation connecting two sequences. Neutral networks an be though of as high, flat plateaus in a [[fitness landscape]]. During [[Neutral theory of molecular evolution|neutral evolution]], genes can randomly move through neutral networks and traverse regions of [[sequence space (evolution)|sequence space]] which may have consequences for [[robustness (evolution)|robustness]] and [[evolvability]]. |
||
| Line 16: | Line 14: | ||
[[File:Neutral network.png|thumb|400px|Each circle represents a functional gene variant and lines represents point mutations between them. Light grid-regions have low [[fitness (biology)|fitness]], dark regions have high fitness. ('''a''') White circles have few neutral neighbours, black circles have many. Light grid-regions contain no circles because those sequences have low fitness. ('''b''') Within a neutral network, the population is predicted to evolve towards the centre and away from ‘fitness cliffs’ (dark arrows).]] |
[[File:Neutral network.png|thumb|400px|Each circle represents a functional gene variant and lines represents point mutations between them. Light grid-regions have low [[fitness (biology)|fitness]], dark regions have high fitness. ('''a''') White circles have few neutral neighbours, black circles have many. Light grid-regions contain no circles because those sequences have low fitness. ('''b''') Within a neutral network, the population is predicted to evolve towards the centre and away from ‘fitness cliffs’ (dark arrows).]] |
||
The more neutral neighbours a sequence has, the more [[robustness (evolution)|robust to mutations]] it is since mutations are more likely to simply neutrally convert it into an equally functional sequence.<ref>{{cite journal|last=van Nimwegen|first=E|coauthors=Crutchfield, JP; Huynen, M|title=Neutral evolution of mutational robustness.|journal=Proceedings of the National Academy of Sciences of the United States of America|date=1999 Aug 17|volume=96|issue=17|pages=9716–20|pmid=10449760}}</ref> Indeed, if there are large differences between the number of neutral neighbours of different sequences within a neutral network, the population is predicted to evolve towards these robust sequences. This is sometimes called circum-neutrality and represents the movement of populations away from cliffs in the [[fitness landscape]].<ref>{{cite journal|last=Proulx|first=SR|coauthors=Adler, FR|title=The standard of neutrality: still flapping in the breeze?|journal=Journal of evolutionary biology|date=2010 Jul|volume=23|issue=7|pages=1339–50|pmid=20492093}}</ref> |
The more neutral neighbours a sequence has, the more [[robustness (evolution)|robust to mutations]] it is since mutations are more likely to simply neutrally convert it into an equally functional sequence.<ref>{{cite journal|last=van Nimwegen|first=E|coauthors=Crutchfield, JP; Huynen, M|title=Neutral evolution of mutational robustness.|journal=Proceedings of the National Academy of Sciences of the United States of America|date=1999 Aug 17|volume=96|issue=17|pages=9716–20|pmid=10449760}}</ref> Indeed, if there are large differences between the number of neutral neighbours of different sequences within a neutral network, the population is predicted to evolve towards these robust sequences. This is sometimes called circum-neutrality and represents the movement of populations away from cliffs in the [[fitness landscape]].<ref>{{cite journal|last=Proulx|first=SR|coauthors=Adler, FR|title=The standard of neutrality: still flapping in the breeze?|journal=Journal of evolutionary biology|date=2010 Jul|volume=23|issue=7|pages=1339–50|pmid=20492093}}</ref> |
||
In addition to in silico models,<ref name="van Nimwegen">{{cite journal | author = van Nimwegen E.,Crutchfield J. P., Huynen M.| title = Neutral evolution of mutational robustness | journal = PNAS | year = 1999| volume = 96 | issue = 17 | pages = 9716–9720}}</ref> these processes are beginning to be confirmed by [[experimental evolution]] of [[cytochrome P450]]s<ref name="Bloom 29">{{cite journal|last=Bloom|first=JD|coauthors=Lu, Z; Chen, D; Raval, A; Venturelli, OS; Arnold, FH|title=Evolution favors protein mutational robustness in sufficiently large populations.|journal=BMC biology|date=2007 Jul 17|volume=5|pages=29|pmid=17640347}}</ref> and [[B-lactamase]].<ref name="Bershtein 2008 1029–1044">{{cite journal|last=Bershtein|first=Shimon|coauthors=Goldin, Korina; Tawfik, Dan S.|title=Intense Neutral Drifts Yield Robust and Evolvable Consensus Proteins|journal=Journal of Molecular Biology|year=2008|month=June|volume=379|issue=5|pages=1029–1044|doi=10.1016/j.jmb.2008.04.024}}</ref> |
|||
===Neutral networks and evolvability=== |
===Neutral networks and evolvability=== |
||
| Line 22: | Line 22: | ||
Interest in the interplay between [[genetic drift]] and selection has been around since the 1930s when the shifting-balance theory proposed that in some situations, genetic drift could facilitate later adaptive evolution.<ref>{{cite journal|last=Wright|first=Sewel|title=The roles of mutation, inbreeding, crossbreeding and selection in evolution|journal=Proceedings of the sixth international congress of genetics|year=1932|pages=356–366}}</ref> Although the specifics of the theory were largely discredited,<ref>{{cite journal|last=Coyne|first=JA|coauthors=Barton NH, Turelli M|title=Perspective: a critique of Sewall Wright's shifting balance theory of evolution|journal=Evolution|year=1997|volume=51|issue=3|pages=643–671}}</ref> it drew attention to the possibility that drift could generate cryptic variation that, though neutral to current function, may affect selection for new functions ([[evolvability]]).<ref>{{cite journal|last=Davies|first=E. K.|title=High Frequency of Cryptic Deleterious Mutations in Caenorhabditis elegans|journal=Science|date=10 September 1999|volume=285|issue=5434|pages=1748–1751|doi=10.1126/science.285.5434.1748}}</ref> |
Interest in the interplay between [[genetic drift]] and selection has been around since the 1930s when the shifting-balance theory proposed that in some situations, genetic drift could facilitate later adaptive evolution.<ref>{{cite journal|last=Wright|first=Sewel|title=The roles of mutation, inbreeding, crossbreeding and selection in evolution|journal=Proceedings of the sixth international congress of genetics|year=1932|pages=356–366}}</ref> Although the specifics of the theory were largely discredited,<ref>{{cite journal|last=Coyne|first=JA|coauthors=Barton NH, Turelli M|title=Perspective: a critique of Sewall Wright's shifting balance theory of evolution|journal=Evolution|year=1997|volume=51|issue=3|pages=643–671}}</ref> it drew attention to the possibility that drift could generate cryptic variation that, though neutral to current function, may affect selection for new functions ([[evolvability]]).<ref>{{cite journal|last=Davies|first=E. K.|title=High Frequency of Cryptic Deleterious Mutations in Caenorhabditis elegans|journal=Science|date=10 September 1999|volume=285|issue=5434|pages=1748–1751|doi=10.1126/science.285.5434.1748}}</ref> |
||
By definition, all genes in a neutral network have equivalent function, however some may exhibit [[promiscuous activities]] which could serve as starting points for adaptive evolution towards new functions.<ref>{{cite journal|last=Masel|first=J|title=Cryptic genetic variation is enriched for potential adaptations.|journal=Genetics|date=2006 Mar|volume=172|issue=3|pages=1985–91|pmid=16387877}}</ref><ref>{{cite journal|last=Hayden|first=EJ|coauthors=Ferrada, E; Wagner, A|title=Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme.|journal=Nature|date=2011 Jun 2|volume=474|issue=7349|pages=92–5|pmid=21637259}}</ref> In terms of [[sequence space (evolution)|sequence space]], current theories predict that if the neutral networks for two different activities overlap, a neutrally evolving population may diffuse to regions of the neutral network of the first activity that allow it to access the second. This would only be the case when the distance between activities is smaller than the distance that a neutrally evolving population can cover. The degree of interpenetration of the two networks will determine how common cryptic variation for the promiscuous activity is in sequence space.<ref>{{cite book|last=Wagner|first=Andreas|title=The origins of evolutionary innovations : a theory of transformative change in living systems|publisher=Oxford University Press|location=Oxford [etc.]|isbn=0199692599}}</ref> |
By definition, all genes in a neutral network have equivalent function, however some may exhibit [[promiscuous activities]] which could serve as starting points for adaptive evolution towards new functions.<ref>{{cite journal|last=Masel|first=J|title=Cryptic genetic variation is enriched for potential adaptations.|journal=Genetics|date=2006 Mar|volume=172|issue=3|pages=1985–91|pmid=16387877}}</ref><ref>{{cite journal|last=Hayden|first=EJ|coauthors=Ferrada, E; Wagner, A|title=Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme.|journal=Nature|date=2011 Jun 2|volume=474|issue=7349|pages=92–5|pmid=21637259}}</ref> In terms of [[sequence space (evolution)|sequence space]], current theories predict that if the neutral networks for two different activities overlap, a neutrally evolving population may diffuse to regions of the neutral network of the first activity that allow it to access the second.<ref>{{cite journal|last=Bornberg-Bauer|first=E|coauthors=Huylmans, AK; Sikosek, T|title=How do new proteins arise?|journal=Current opinion in structural biology|date=2010 Jun|volume=20|issue=3|pages=390-6|pmid=20347587}}</ref> This would only be the case when the distance between activities is smaller than the distance that a neutrally evolving population can cover. The degree of interpenetration of the two networks will determine how common cryptic variation for the promiscuous activity is in sequence space.<ref>{{cite book|last=Wagner|first=Andreas|title=The origins of evolutionary innovations : a theory of transformative change in living systems|publisher=Oxford University Press|location=Oxford [etc.]|isbn=0199692599}}</ref> |
||
==References== |
==References== |
||
Revision as of 00:53, 15 January 2014
A Neutral network is a set of genes all related by point mutations that have equivalent function or fitness.[1] Each node represents a gene sequence and each line represents the mutation connecting two sequences. Neutral networks an be though of as high, flat plateaus in a fitness landscape. During neutral evolution, genes can randomly move through neutral networks and traverse regions of sequence space which may have consequences for robustness and evolvability.
Genetic and molecular causes
Neutral networks exist in fitness landscapes since proteins are robust to mutations. This leads to extended networks of genes of equivalent function, linked by neutral mutations.[2][3] Proteins are resistant to mutations because many sequences can fold into highly similar structural folds.[4] A protein adopts a limited ensemble of native conformations because those conformers have lower energy than unfolded and mis-folded states (ΔΔG of folding).[5][6] This is achieved by a distributed, internal network of cooperative interactions (hydrophobic, polar and covalent).[7] Protein structural robustness results from few single mutations being sufficiently disruptive to compromise function. Proteins have also evolved to avoid aggregation[8] as partially folded proteins can combine to form large, repeating, insoluble protein fibrils and masses.[9] There is evidence that proteins show negative design features to reduce the exposure of aggregation-prone beta-sheet motifs in their structures.[10] Additionally, there is some evidence that the genetic code itself may be optimised such that most point mutations lead to similar amino acids (conservative).[11][12] Together these factors create a distribution of fitness effects of mutations that contains a high proportion of neutral and nearly-neutral mutations.[13]
Neutral networks and evolution
Neutral networks are a subset of the sequences in sequence space that have equivalent function, and so form a wide, flat plateau in a fitness landscape. Neutral evolution can therefore be visualised as a population diffusing from one set of sequence nodes, through the neutral network, to another cluster of sequence nodes. Since the majority of evolution is thought to be neutral,[14][15] a large proportion of gene change is the movement though expansive neutral networks.
Neutral networks and robustness
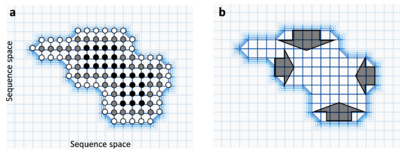
The more neutral neighbours a sequence has, the more robust to mutations it is since mutations are more likely to simply neutrally convert it into an equally functional sequence.[16] Indeed, if there are large differences between the number of neutral neighbours of different sequences within a neutral network, the population is predicted to evolve towards these robust sequences. This is sometimes called circum-neutrality and represents the movement of populations away from cliffs in the fitness landscape.[17]
In addition to in silico models,[18] these processes are beginning to be confirmed by experimental evolution of cytochrome P450s[19] and B-lactamase.[20]
Neutral networks and evolvability
Interest in the interplay between genetic drift and selection has been around since the 1930s when the shifting-balance theory proposed that in some situations, genetic drift could facilitate later adaptive evolution.[21] Although the specifics of the theory were largely discredited,[22] it drew attention to the possibility that drift could generate cryptic variation that, though neutral to current function, may affect selection for new functions (evolvability).[23]
By definition, all genes in a neutral network have equivalent function, however some may exhibit promiscuous activities which could serve as starting points for adaptive evolution towards new functions.[24][25] In terms of sequence space, current theories predict that if the neutral networks for two different activities overlap, a neutrally evolving population may diffuse to regions of the neutral network of the first activity that allow it to access the second.[26] This would only be the case when the distance between activities is smaller than the distance that a neutrally evolving population can cover. The degree of interpenetration of the two networks will determine how common cryptic variation for the promiscuous activity is in sequence space.[27]
References
- ^ van Nimwegen, E (1999 Aug 17). "Neutral evolution of mutational robustness". Proceedings of the National Academy of Sciences of the United States of America. 96 (17): 9716–20. PMID 10449760.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Taverna, DM (2002 Jan 18). "Why are proteins so robust to site mutations?". Journal of molecular biology. 315 (3): 479–84. PMID 11786027.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Tokuriki, N (2009 Oct). "Stability effects of mutations and protein evolvability". Current opinion in structural biology. 19 (5): 596–604. PMID 19765975.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Meyerguz, L (2007 Jul 10). "The network of sequence flow between protein structures". Proceedings of the National Academy of Sciences of the United States of America. 104 (28): 11627–32. PMID 17596339.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Karplus, M (2011 Jun 17). "Behind the folding funnel diagram". Nature chemical biology. 7 (7): 401–4. PMID 21685880.
{{cite journal}}: Check date values in:|date=(help) - ^ Tokuriki, N (2007 Jun 22). "The stability effects of protein mutations appear to be universally distributed". Journal of molecular biology. 369 (5): 1318–32. PMID 17482644.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Shakhnovich, BE (2005 Mar). "Protein structure and evolutionary history determine sequence space topology". Genome research. 15 (3): 385–92. PMID 15741509.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Monsellier, E (2007 Aug). "Prevention of amyloid-like aggregation as a driving force of protein evolution". EMBO reports. 8 (8): 737–42. PMID 17668004.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Fink, AL (1998). "Protein aggregation: folding aggregates, inclusion bodies and amyloid". Folding & design. 3 (1): R9-23. PMID 9502314.
- ^ Richardson, JS (2002 Mar 5). "Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation". Proceedings of the National Academy of Sciences of the United States of America. 99 (5): 2754–9. PMID 11880627.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Müller, MM (2013). "Directed evolution of a model primordial enzyme provides insights into the development of the genetic code". PLoS genetics. 9 (1): e1003187. PMID 23300488.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Firnberg, E (2013 Aug). "The genetic code constrains yet facilitates Darwinian evolution". Nucleic acids research. 41 (15): 7420–8. PMID 23754851.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Hietpas, RT (2011 May 10). "Experimental illumination of a fitness landscape". Proceedings of the National Academy of Sciences of the United States of America. 108 (19): 7896–901. PMID 21464309.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Kimura, Motoo. (1983). The neutral theory of molecular evolution. Cambridge
- ^ Kimura M. (1968). Evolutionary Rate at the Molecular Level. Nature 217:624-6.
- ^ van Nimwegen, E (1999 Aug 17). "Neutral evolution of mutational robustness". Proceedings of the National Academy of Sciences of the United States of America. 96 (17): 9716–20. PMID 10449760.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Proulx, SR (2010 Jul). "The standard of neutrality: still flapping in the breeze?". Journal of evolutionary biology. 23 (7): 1339–50. PMID 20492093.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ van Nimwegen E.,Crutchfield J. P., Huynen M. (1999). "Neutral evolution of mutational robustness". PNAS. 96 (17): 9716–9720.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bloom, JD (2007 Jul 17). "Evolution favors protein mutational robustness in sufficiently large populations". BMC biology. 5: 29. PMID 17640347.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Bershtein, Shimon (2008). "Intense Neutral Drifts Yield Robust and Evolvable Consensus Proteins". Journal of Molecular Biology. 379 (5): 1029–1044. doi:10.1016/j.jmb.2008.04.024.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Wright, Sewel (1932). "The roles of mutation, inbreeding, crossbreeding and selection in evolution". Proceedings of the sixth international congress of genetics: 356–366.
- ^ Coyne, JA (1997). "Perspective: a critique of Sewall Wright's shifting balance theory of evolution". Evolution. 51 (3): 643–671.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Davies, E. K. (10 September 1999). "High Frequency of Cryptic Deleterious Mutations in Caenorhabditis elegans". Science. 285 (5434): 1748–1751. doi:10.1126/science.285.5434.1748.
- ^ Masel, J (2006 Mar). "Cryptic genetic variation is enriched for potential adaptations". Genetics. 172 (3): 1985–91. PMID 16387877.
{{cite journal}}: Check date values in:|date=(help) - ^ Hayden, EJ (2011 Jun 2). "Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme". Nature. 474 (7349): 92–5. PMID 21637259.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Bornberg-Bauer, E (2010 Jun). "How do new proteins arise?". Current opinion in structural biology. 20 (3): 390–6. PMID 20347587.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Wagner, Andreas. The origins of evolutionary innovations : a theory of transformative change in living systems. Oxford [etc.]: Oxford University Press. ISBN 0199692599.
