DNA demethylation: Difference between revisions
→External links: too specific to the article. |
No edit summary |
||
| Line 53: | Line 53: | ||
Altered protein expression in neurons, (likely initiated by 8-oxo-dG-dependent demethylation of CpG sites in gene promoters within neuron DNA), is central to memory formation.<ref name="pmid20975755">{{cite journal |vauthors=Day JJ, Sweatt JD |title=DNA methylation and memory formation |journal=Nat. Neurosci. |volume=13 |issue=11 |pages=1319–23 |date=November 2010 |pmid=20975755 |pmc=3130618 |doi=10.1038/nn.2666 |url=}}</ref> |
Altered protein expression in neurons, (likely initiated by 8-oxo-dG-dependent demethylation of CpG sites in gene promoters within neuron DNA), is central to memory formation.<ref name="pmid20975755">{{cite journal |vauthors=Day JJ, Sweatt JD |title=DNA methylation and memory formation |journal=Nat. Neurosci. |volume=13 |issue=11 |pages=1319–23 |date=November 2010 |pmid=20975755 |pmc=3130618 |doi=10.1038/nn.2666 |url=}}</ref> |
||
=== |
===Demethylation after exercise=== |
||
Physical exercise has well established beneficial effects on learning and memory (see [[Neurobiological effects of physical exercise]]). ''[[Brain-derived neurotrophic factor|BDNF]]'' is a particularly important regulator of learning and memory.<ref name="pmid23587647">{{cite journal |vauthors=Karpova NN |title=Role of BDNF epigenetics in activity-dependent neuronal plasticity |journal=Neuropharmacology |volume=76 Pt C |issue= |pages=709–18 |date=January 2014 |pmid=23587647 |doi=10.1016/j.neuropharm.2013.04.002 |url=}}</ref> As reviewed by Fernandes et al.,<ref name=Fernandes>{{cite journal |vauthors=Fernandes J, Arida RM, Gomez-Pinilla F |title=Physical exercise as an epigenetic modulator of brain plasticity and cognition |journal=Neurosci Biobehav Rev |volume=80 |issue= |pages=443–456 |date=September 2017 |pmid=28666827 |pmc=5705447 |doi=10.1016/j.neubiorev.2017.06.012 |url=}}</ref> in rats, exercise enhances the [[hippocampus]] expression of the gene ''[[Brain-derived neurotrophic factor|Bdnf]]'', which has an essential role in memory formation. [[Downregulation and upregulation|Enhanced expression]] of ''Bdnf'' occurs through demethylation of its [[CpG site#CpG island promoter|CpG island promoter]] at [[Brain-derived neurotrophic factor#expression|exon IV]]<ref name=Fernandes /> and this demethylation depends on steps illustrated in the two figures.<ref name="pmid29875631" /> |
Physical exercise has well established beneficial effects on learning and memory (see [[Neurobiological effects of physical exercise]]). ''[[Brain-derived neurotrophic factor|BDNF]]'' is a particularly important regulator of learning and memory.<ref name="pmid23587647">{{cite journal |vauthors=Karpova NN |title=Role of BDNF epigenetics in activity-dependent neuronal plasticity |journal=Neuropharmacology |volume=76 Pt C |issue= |pages=709–18 |date=January 2014 |pmid=23587647 |doi=10.1016/j.neuropharm.2013.04.002 |url=}}</ref> As reviewed by Fernandes et al.,<ref name=Fernandes>{{cite journal |vauthors=Fernandes J, Arida RM, Gomez-Pinilla F |title=Physical exercise as an epigenetic modulator of brain plasticity and cognition |journal=Neurosci Biobehav Rev |volume=80 |issue= |pages=443–456 |date=September 2017 |pmid=28666827 |pmc=5705447 |doi=10.1016/j.neubiorev.2017.06.012 |url=}}</ref> in rats, exercise enhances the [[hippocampus]] expression of the gene ''[[Brain-derived neurotrophic factor|Bdnf]]'', which has an essential role in memory formation. [[Downregulation and upregulation|Enhanced expression]] of ''Bdnf'' occurs through demethylation of its [[CpG site#CpG island promoter|CpG island promoter]] at [[Brain-derived neurotrophic factor#expression|exon IV]]<ref name=Fernandes /> and this demethylation depends on steps illustrated in the two figures.<ref name="pmid29875631" /> |
||
===Demethylation after exposure to traffic related air pollution=== |
|||
In a panel of healthy adults, negative associations were found between total DNA methylation and exposure to traffic related air pollution. DNA methylation levels were associated both with recent and chronic exposure to Black Carbon as well as benzene. <ref name="louwies">{{cite journal |vauthors=Louwies T |title=DNA hypomethylation in association with internal and external markers of traffic exposure in a panel of healthy adults |journal=Air Quality, Atmosphere & Health |volume=11 |issue=6 |pages=673–681 |date= 2018 |doi=https://doi.org/10.1007/s11869-018-0574-4}}</ref> |
|||
==Peripheral sensory neuron regeneration== |
==Peripheral sensory neuron regeneration== |
||
Revision as of 07:36, 7 November 2019
This article has multiple issues. Please help improve it or discuss these issues on the talk page. (Learn how and when to remove these template messages)
|
DNA demethylation is the process of removal of a methyl group from nucleotides in DNA. Both DNA demethylation and methylation play important roles in mammalian development and differentiation, as well as in cognition and neuroregeneration (after injury to peripheral nerves in mammals). DNA methylation on cytosine at CpG sites on a gene promoter leads to the silencing of gene expression, while DNA demethylation of a gene promoter is linked to transcriptional activation and gene expression. These are called epigenetic changes. The DNA methylation status that controls gene expression is copied during DNA replication and is transmitted to daughter cells along with the DNA sequence. This area is currently being investigated for its role in disease progression and for potential treatments, such as cancer therapy.[1]
Passive and active demethylation
DNA demethylation can occur through passive or active mechanisms. The passive process takes place in the absence of methylation of newly synthesised DNA strands by DNMT1 during several replication rounds (for example, upon 5-Azacytidine treatment), leading to dilution of the methylation signal. Active DNA demethylation is mediated by multiple enzymes and can occur independent of DNA replication.
Examples DNA Demethylation
All the cases of DNA demethylation can be classified as global (genome wide) or locus-specific (when just specific sequences are demethylated). The genome-wide DNA demethylation occurs:
- In mammals:
- In the male pronucleus of zygote immediately after fertilization;
- In mouse primordial germ cells (PGCs) between E8.5-11.5 day old embryos;[2]
- Possibly in amphibia - during midblastula transition.
Examples of specific DNA demethylation:
- Genomic imprinting during plant reproduction;
- Electroconvulsive stimulation-induced demethylation of neurotrophic factor genes in dentate gyrus neurons in the mouse brain.[3][4]
Possible mechanisms of active DNA demethylation
There are several proposed hypothetical mechanisms of active DNA demethylation:
A Direct removal of 5-methylcytosine
- Direct removal of methyl group. This process has quite low thermodynamic probability.
- Removal of methylated bases (either by direct removal of methylcytosine, or through cytosine deamination followed by removal of thymine from thymine/guanosine mismatch), followed by insertion of unmethylated one using base excision repair machinery (BER).
- Removal of entire DNA patch and following filling it with new nucleotides by nucleotide excision repair (NER) or mismatch repair (MMR).[5]
B Removal of 5-methylcytosine via further modified cytosine bases
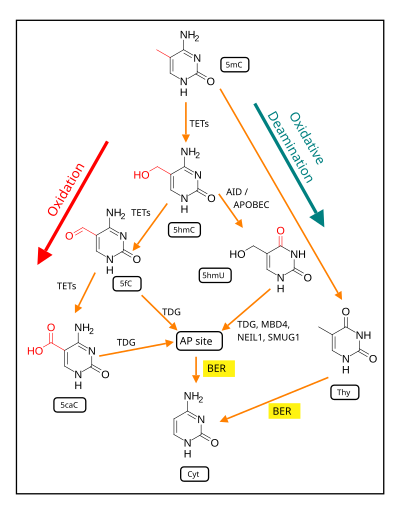
Oxidation of the methyl group generates 5-Hydroxymethylcytosine. Several mechanisms have been proposed to mediate demethylation of 5-hydroxymethylcytosines.[6][7] This base can be either deaminated by AID/Apobec enzymes to give 5-Hydroxymethyluracil.[4] Alternatively, TET enzymes can further oxidize 5-hydroxymethylcytosine to 5-Formylcytosine and 5-Carboxylcytosine.[8][9][10]
- Both the deamination and the oxidation products have been shown to be repaired by TDG, a glycosylase which is involved in base excision repair.[9][11][12] A base excision mediated demethylation mechanism would yield double strand breaks if it occurs on large scale in CpG islands.
- The carboxyl and formyl groups of 5-Formylcytosine and 5-Carboxylcytosine could be enzymatically removed without excision of the base.[6][7][8][10] Precedent for similar reactions is found in biosynthetic pathways.
DNA hydroxymethylation
DNA hydroxymethylation has been proposed to act as a specific epigenetic mark opposing DNA methylation, rather than a passive intermediate in the de-methylation pathway. DNA hydroxymethylation in vivo is sometimes associated with labile nucleosomes, which are more easy to disassemble and to be out-competed by transcription factors during cell development.[13] Hydroxymethylation has been associated with pluripotency of stem cells. Furthermore, changes in hydroxymethylation have been associated with cancer.[14]
Cognition
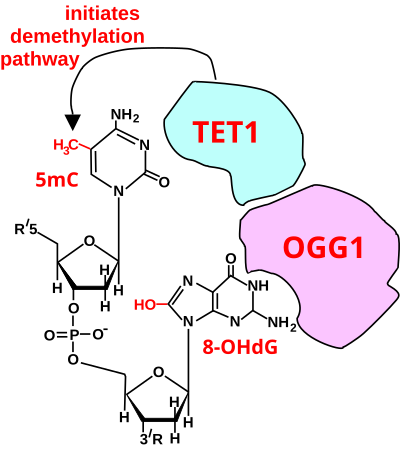
In mammals, DNA methyltransferases (which add methyl groups to DNA bases) exhibit a strong sequence preference for cytosines within the particular DNA sequence cytosine-phosphate-guanine (CpG sites).[17] Methylation of cytosines occurs at 60–90% of CpG sites depending on the tissue type.[18] In the mammalian brain, ~62% of CpGs are methylated.[18] Methylation of CpG sites tends to stably silence genes.[19]
Active DNA methylation and demethylation is required for the cognition process of memory formation and maintenance.[20] In rats, contextual fear conditioning can trigger lifelong memory for the event with a single trial, and methylation changes appear to be correlated with triggering particularly long-lived memories.[20] With contextual fear conditioning, after 24 hours, DNA isolated from the rat brain hippocampus region had 2097 differentially methylated genes, with a portion being demethylated.[20] Similar results in the hippocampus were obtained with contextual fear conditioning in mice.[21] As shown with the rats, 9.2% of the genes in the rat hippocampus neurons are differentially methylated 24 hours after contextual fear conditioning. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex of mice four weeks after contextual fear conditioning. Thus, where there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as 4 weeks later.
The process of demethylation is illustrated in the two figures shown in this section. First, the guanine in the CpG site is oxidized to form 8-oxo-dG (or its tautomer 8-OHdG) (see first figure).[15] TET1 is a key enzyme involved in demethylating 5mCpG. However, TET1 is only able to act on 5mCpG if the guanine has first been oxidized (presumably by an ROS) to form 8-OHdG, resulting in a 5mCp-8-OHdG dinucleotide (see first figure).[15] After formation of 5mCp-8-OHdG, the base excision repair enzyme OGG1 binds to the 8-OHdG lesion without immediate excision. Adherence of OGG1 to the 5mCp-8-OHdG site recruits TET1, allowing TET1 to oxidize the 5mC adjacent to 8-OHdG, as shown in the second figure. As reviewed by Bayraktar and Kreutz,[16] in the brain, further reactions in DNA demethylation are primarily dependent upon TET enzymes in the steps indicated in the second figure. The formation of the final product, unmethylated cytosine, depends on base excision repair (BER) as the terminal step.
Altered protein expression in neurons, (likely initiated by 8-oxo-dG-dependent demethylation of CpG sites in gene promoters within neuron DNA), is central to memory formation.[22]
Demethylation after exercise
Physical exercise has well established beneficial effects on learning and memory (see Neurobiological effects of physical exercise). BDNF is a particularly important regulator of learning and memory.[23] As reviewed by Fernandes et al.,[24] in rats, exercise enhances the hippocampus expression of the gene Bdnf, which has an essential role in memory formation. Enhanced expression of Bdnf occurs through demethylation of its CpG island promoter at exon IV[24] and this demethylation depends on steps illustrated in the two figures.[16]
In a panel of healthy adults, negative associations were found between total DNA methylation and exposure to traffic related air pollution. DNA methylation levels were associated both with recent and chronic exposure to Black Carbon as well as benzene. [25]
Peripheral sensory neuron regeneration
After injury, neurons in the adult peripheral nervous system can switch from a dormant state with little axonal growth to robust axon regeneration. DNA demethylation in mature mammalian neurons removes barriers to axonal regeneration.[26] This demethylation, in regenerating mouse peripheral neurons, depends upon TET3 to generate 5-hydroxymethylcytosine (5hmC) in DNA.[26][27] 5hmC was altered in a large set of regeneration-associated genes (RAGs), including well-known RAGs such as Atf3, Bdnf, and Smad1, that regulate the axon growth potential of neurons.[27]
References
- ^ "DNA demethylation". Epigentek. Retrieved 1 August 2018.
- ^ Hackett, JA; Sengupta, R; Zylicz, JJ; Murakami, K; Lee, C; Down, T; Surani, MA (2012-12-06). "Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine". Science. 339 (6118): 448–52. doi:10.1126/science.1229277. PMC 3847602. PMID 23223451.
- ^ Ma, DK; Jang, MH; Guo, JU; Kitabatake, Y; Chang, ML; Pow-Anpongkul, N; Flavell, RA; Lu, B; Ming, GL; Song, H (2009-02-20). "Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis". Science. 323 (5917): 1074–7. doi:10.1126/science.1166859. PMC 2726986. PMID 19119186.
- ^ a b Guo, JU; Su, Y; Zhong, C; Ming, GL; Song, H (2011-04-29). "Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain". Cell. 145 (3): 423–34. doi:10.1016/j.cell.2011.03.022. PMC 3088758. PMID 21496894.
- ^ Grin, I; Ishchenko, AA (May 2016). "An interplay of the base excision repair and mismatch repair pathways in active DNA demethylation". Nucleic Acids Res. 44 (8): 3713–27. doi:10.1093/nar/gkw059. PMC 4856981. PMID 26843430.
- ^ a b Wu, SC; Zhang, Y (Sep 2010). "Active DNA demethylation: many roads lead to Rome". Nature Reviews Molecular Cell Biology. 11 (9): 607–20. doi:10.1038/nrm2950. PMC 3711520. PMID 20683471.
- ^ a b Globisch, Daniel; Münzel, Martin; Müller, Markus; Michalakis, Stylianos; Wagner, Mirko; Koch, Susanne; Brückl, Tobias; Biel, Martin; Carell, Thomas (23 December 2010). Croft, Anna Kristina (ed.). "Tissue Distribution of 5-Hydroxymethylcytosine and Search for Active Demethylation Intermediates". PLoS ONE. 5 (12): e15367. doi:10.1371/journal.pone.0015367. PMC 3009720. PMID 21203455.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Pfaffeneder, Toni; Hackner, Benjamin; Truss, Matthias; Münzel, Martin; Müller, Markus; Deiml, Christian A.; Hagemeier, Christian; Carell, Thomas (30 June 2011). "The Discovery of 5-Formylcytosine in Embryonic Stem Cell DNA". Angew. Chem. Int. Ed. 50 (31): 7008–7012. doi:10.1002/anie.201103899. PMID 21721093.
- ^ a b He, YF; Li, BZ; Li, Z; Liu, P; Wang, Y; Tang, Q; Ding, J; Jia, Y; Chen, Z; Li, L; Sun, Y; Li X; Dai, Q; Song, CX; Zhang, K; He, C; Xu, GL (4 August 2011). "Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA". Science. 333 (6047): 1303–1307. doi:10.1126/science.1210944. PMC 3462231. PMID 21817016.
- ^ a b Ito, S; Li, S; Dai, Q; Wu, SC; Collins, SB; Swenberg, JA; He, C; Zhang, Y (21 July 2011). "Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine". Science. 333 (6047): 1300–1303. doi:10.1126/science.1210597. PMC 3495246. PMID 21778364.
- ^ Maiti, A; Drohat, AC (23 August 2011). "Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine". J. Biol. Chem. 286 (41): 35334–8. doi:10.1074/jbc.C111.284620. PMC 3195571. PMID 21862836.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Cannon, SV; Cummings, GW; Teebor, GW (1988). "5-Hydroxymethylcytosine DNA Glycosylase Activity in Mammalian Tissue". Biochem. Biophys. Res. Commun. 151 (3): 1173–1179. doi:10.1016/S0006-291X(88)80489-3. PMID 3355548.
- ^ Teif, Vladimir; Beshnova, Daria A.; Vainshtein, Yevhen; Marth, Caroline; Mallm, Jan-Philipp; Höfer, Thomas; Rippe, Karsten (8 May 2014). "Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development". Genome Research. 24 (8): 1285–1295. doi:10.1101/gr.164418.113. PMC 4120082. PMID 24812327.
- ^ http://www.ks.uiuc.edu/Research/methylation/
- ^ a b c Zhou X, Zhuang Z, Wang W, He L, Wu H, Cao Y, Pan F, Zhao J, Hu Z, Sekhar C, Guo Z (September 2016). "OGG1 is essential in oxidative stress induced DNA demethylation". Cell. Signal. 28 (9): 1163–71. doi:10.1016/j.cellsig.2016.05.021. PMID 27251462.
- ^ a b c Bayraktar G, Kreutz MR (2018). "The Role of Activity-Dependent DNA Demethylation in the Adult Brain and in Neurological Disorders". Front Mol Neurosci. 11: 169. doi:10.3389/fnmol.2018.00169. PMC 5975432. PMID 29875631.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Ziller MJ, Müller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, Gnirke A, Meissner A (December 2011). "Genomic distribution and inter-sample variation of non-CpG methylation across human cell types". PLoS Genet. 7 (12): e1002389. doi:10.1371/journal.pgen.1002389. PMC 3234221. PMID 22174693.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Fasolino M, Zhou Z (May 2017). "The Crucial Role of DNA Methylation and MeCP2 in Neuronal Function". Genes (Basel). 8 (5). doi:10.3390/genes8050141. PMC 5448015. PMID 28505093.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Bird A (January 2002). "DNA methylation patterns and epigenetic memory". Genes Dev. 16 (1): 6–21. doi:10.1101/gad.947102. PMID 11782440.
- ^ a b c Duke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD (July 2017). "Experience-dependent epigenomic reorganization in the hippocampus". Learn. Mem. 24 (7): 278–288. doi:10.1101/lm.045112.117. PMC 5473107. PMID 28620075.
- ^ Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, Centeno TP, van Bebber F, Capece V, Garcia Vizcaino JC, Schuetz AL, Burkhardt S, Benito E, Navarro Sala M, Javan SB, Haass C, Schmid B, Fischer A, Bonn S (January 2016). "DNA methylation changes in plasticity genes accompany the formation and maintenance of memory". Nat. Neurosci. 19 (1): 102–10. doi:10.1038/nn.4194. PMC 4700510. PMID 26656643.
- ^ Day JJ, Sweatt JD (November 2010). "DNA methylation and memory formation". Nat. Neurosci. 13 (11): 1319–23. doi:10.1038/nn.2666. PMC 3130618. PMID 20975755.
- ^ Karpova NN (January 2014). "Role of BDNF epigenetics in activity-dependent neuronal plasticity". Neuropharmacology. 76 Pt C: 709–18. doi:10.1016/j.neuropharm.2013.04.002. PMID 23587647.
- ^ a b Fernandes J, Arida RM, Gomez-Pinilla F (September 2017). "Physical exercise as an epigenetic modulator of brain plasticity and cognition". Neurosci Biobehav Rev. 80: 443–456. doi:10.1016/j.neubiorev.2017.06.012. PMC 5705447. PMID 28666827.
- ^ Louwies T (2018). "DNA hypomethylation in association with internal and external markers of traffic exposure in a panel of healthy adults". Air Quality, Atmosphere & Health. 11 (6): 673–681. doi:https://doi.org/10.1007/s11869-018-0574-4.
{{cite journal}}: Check|doi=value (help); External link in|doi= - ^ a b Weng YL, An R, Cassin J, Joseph J, Mi R, Wang C, Zhong C, Jin SG, Pfeifer GP, Bellacosa A, Dong X, Hoke A, He Z, Song H, Ming GL (April 2017). "An Intrinsic Epigenetic Barrier for Functional Axon Regeneration". Neuron. 94 (2): 337–346.e6. doi:10.1016/j.neuron.2017.03.034. PMC 6007997. PMID 28426967.
- ^ a b Loh YE, Koemeter-Cox A, Finelli MJ, Shen L, Friedel RH, Zou H (February 2017). "Comprehensive mapping of 5-hydroxymethylcytosine epigenetic dynamics in axon regeneration". Epigenetics. 12 (2): 77–92. doi:10.1080/15592294.2016.1264560. PMC 5330438. PMID 27918235.
