Deep homology

In evolutionary developmental biology, the concept of deep homology is used to describe cases where growth and differentiation processes are governed by genetic mechanisms that are homologous and deeply conserved across a wide range of species.
History
In 1822, the French zoologist Étienne Geoffroy Saint-Hilaire dissected a crayfish, discovering that its body is organised like a vertebrate's, but inverted belly to back (dorsoventrally):[1]
I just found that all the soft organs, that is to say, the principal organs of life are found in crustaceans, and so in insects, in the same order, in the same relationships and with the same arrangement as their analogues in the high vertebrate animals ... What was my surprise, and I may add, my admiration, seeing [such] a rule ...[1]
Geoffroy's homology theory was denounced by the leading French zoologist of his day, Georges Cuvier, but in 1994, Geoffroy was shown to be correct.[1] In 1915, Santiago Ramon y Cajal mapped the neural connections of the optic lobes of a fly, finding that these resembled those of vertebrates.[1] In 1978, Edward B. Lewis helped to found evolutionary developmental biology, discovering that homeotic genes regulated embryonic development in fruit flies.[1]
In 1997, the term deep homology first appeared in a paper by Neil Shubin, Cliff Tabin, and Sean B. Carroll, describing the apparent relatedness in genetic regulatory apparatuses which indicated evolutionary similarities in disparate animal features.[2]
A different kind of homology
Whereas ordinary homology is seen in the pattern of structures such as limb bones of mammals that are evidently related, deep homology can apply to groups of animals that have quite dissimilar anatomy: vertebrates (with endoskeletons made of bone and cartilage) and arthropods (with exoskeletons made of chitin) nevertheless have limbs that are constructed using similar recipes or "algorithms".[2][3][4][5]
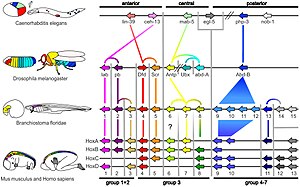
Within the metazoa, homeotic genes control differentiation along major body axes, and pax genes (especially PAX6) help to control the development of the eye and other sensory organs. The deep homology applies across widely separated groups, such as in the eyes of mammals and the structurally quite different compound eyes of insects.[3]
Similarly, hox genes help to form an animal's segmentation pattern. HoxA and HoxD, that regulate finger and toe formation in mice, control the development of ray fins in zebrafish; these structures had until then been considered non-homologous.[6]
There is a possible deep homology among animals that use acoustic communication, such as songbirds and humans, which may share unmutated versions of the FOXP2 gene.[7]
Algorithm
In 2010, a team led by Edward Marcotte developed an algorithm that identifies deeply homologous genetic modules in unicellular organisms, plants, and animals based on phenotypes (such as traits and developmental defects). The technique aligns phenotypes across organisms based on orthology (a type of homology) of genes involved in the phenotypes.[8][9]
References
- ^ a b c d e Held, Lewis I. (February 2017). Deep Homology?: Uncanny Similarities of Humans and Flies Uncovered by Evo-Devo. Cambridge University Press. pp. 2–5. ISBN 978-1316601211.
- ^ a b Shubin, Neil; Tabin, Cliff; Carroll, Sean (1997). "Fossils, genes and the evolution of animal limbs". Nature. 388 (6643). Springer Nature: 639–648. Bibcode:1997Natur.388..639S. doi:10.1038/41710. PMID 9262397. S2CID 2913898.
- ^ a b Carroll, Sean B. (2006). Endless Forms Most Beautiful. Weidenfeld & Nicolson. pp. 28, 66–69. ISBN 0-297-85094-6.
- ^ Gilbert, Scott F. (2000). "Homologous Pathways of Development". Developmental biology (6th ed.). Sunderland, Mass: Sinauer Associates. ISBN 0-87893-243-7.
- ^ Held, Lewis I. (February 2017). Deep Homology?: Uncanny Similarities of Humans and Flies Uncovered by Evo-Devo. Cambridge University Press. pp. viii and throughout. ISBN 978-1316601211.
- ^ Zimmer, Carl (2016-08-17). "From Fins Into Hands: Scientists Discover a Deep Evolutionary Link". The New York Times. Retrieved 21 October 2016.
- ^ Scharff, Petri; Constance, Jane (July 2011). "Evo-Devo, Deep Homology and FoxP2: Implications for the Evolution of Speech and Language". Philos. Trans. R. Soc. B. 366 (1574): 2124–2140. doi:10.1098/rstb.2011.0001. PMC 3130369. PMID 21690130.
- ^ Zimmer, Carl (April 26, 2010). "The Search for Genes Leads to Unexpected Places". The New York Times.
- ^ McGary, K. L.; Park, T. J.; Woods, J. O.; Cha, H. J.; Wallingford, J. B.; Marcotte, E. M. (April 2010). "Systematic discovery of nonobvious human disease models through orthologous phenotypes" (PDF). Proceedings of the National Academy of Sciences. 107 (14): 6544–9. Bibcode:2010PNAS..107.6544M. doi:10.1073/pnas.0910200107. PMC 2851946. PMID 20308572.
See also
- Body plan – Set of morphological features common to members of a phylum of animals
