Penetrance

Penetrance in genetics is the proportion of individuals carrying a particular variant (or allele) of a gene (genotype) that also expresses an associated trait (phenotype). In medical genetics, the penetrance of a disease-causing mutation is the proportion of individuals with the mutation that exhibit clinical symptoms among all individuals with such mutation.[1] For example: If a mutation in the gene responsible for a particular autosomal dominant disorder has 75% penetrance, then 75% of those with the mutation will go on to develop the disease, showing its phenotype, whereas 25% will not.

Penetrance only refers to whether an individual with a specific genotype exhibits any phenotypic signs or symptoms, and is not to be confused with variable expressivity which is to what extent or degree the symptoms for said disease are shown (the expression of the phenotypic trait). Meaning that, even if the same disease-causing mutation affects separate individuals, the expressivity will vary.[1][2] [3]
Degrees of Penetrance[edit]
Complete penetrance[edit]
If 100% of individuals carrying a particular genotype express the associated trait, the genotype is said to show complete penetrance.[1] Neurofibromatosis type 1 (NF1), is an autosomal dominant condition which shows complete penetrance, consequently everyone who inherits the disease-causing variant of this gene will develop some degree of symptoms for NF1.[4]
Reduced penetrance[edit]
The penetrance is said to be reduced if less than 100% of individuals carrying a particular genotype express associated traits, and is likely to be caused by a combination of genetic, environmental and lifestyle factors.[1][3] BRCA1 is an example of a genotype with reduced penetrance. By age 70, the mutation is estimated to have a breast cancer penetrance of around 65% in women. Meaning that about 65% of women carrying the gene will develop breast cancer by the time they turn 70.[5]
- Non-penetrance: Within the category of reduced penetrance, individuals carrying the mutation without displaying any signs or symptoms, are said to have a genotype that is non-penetrant. For the BRCA1 example above, the remaining 35% which never develop breast cancer, are therefore carrying the mutation, but it is non-penetrant. This can lead to healthy, unaffected parents carrying the mutation on to future generations that might be affected.[6]
Factors affecting penetrance[edit]
Many factors such as age, sex, environment, epigenetic modifiers, and modifier genes are linked to penetrance. These factors can help explain why certain individuals with a specific genotype exhibit symptoms or signs of disease, whilst others do not. [1][3]
Age-dependent penetrance[edit]
If clinical signs associated with a specific genotype appear more frequently with increasing age, the penetrance is said to be age dependent. Some diseases are non-penetrant up until a certain age and then the penetrance starts to increase drastically, whilst others exhibit low penetrance at an early age and continue to increase with time. For this reason, many diseases have a different estimated penetrance dependent on the age.[1]
A specific hexanucleotide repeat expansion within the C9orf72 gene said to be a major cause for developing amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) is an example of a genotype with age dependent penetrance. The genotype is said to be non-penetrant until the age of 35, 50% penetrant by the age of 60, and almost completely penetrant by age 80. [1][7]
[edit]
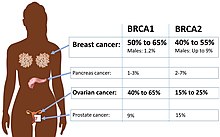
For some mutations, the phenotype is more frequently present in one sex and in rare cases mutations appear completely non-penetrant in a particular gender. This is called gender-related penetrance or sex-dependent penetrance and may be the result of allelic variation, disorders in which the expression of the disease is limited to organs only found in one sex such as testis or ovaries, or sex steroid-responsive genes.[1][3][9] Breast cancer caused by the BRCA2 mutation is an example of a disease with gender-related penetrance. The penetrance is determined to be much higher in women than men. By age 70, around 86% of females in contrast to 6% of males with the same mutation is estimated to develop breast cancer.[9]
In cases where clinical symptoms or the phenotype related to a genetic mutation are present only in one sex, the disorder is said to be sex-limited. Familial male-limited precocious puberty (FMPP) caused by a mutation in the LHCGR gene, is an example of a genotype only penetrant in males. Meaning that males with this particular genotype exhibit symptoms of the disease whilst the same genotype is nonpenetrant in females.[3][9][10]
Genetic modifiers[edit]
Genetic modifiers are genetic variants or mutations able to modify a primary disease-causing variant's phenotypic outcome without being disease causing themselves.[11] For instance, in single gene disorders there is one gene primarily responsible for development of the disease, but modifier genes inherited separately can affect the phenotype. Meaning that the presence of a mutation located on a loci different from the one with the disease-causing mutation, may either hinder manifestation of the phenotype or alter the mutations effects, and thereby influencing the penetrance.[1][3]
Environmental modifiers[edit]
Exposure to environmental and lifestyle factors such as chemicals, diet, alcohol intake, drugs and stress are some of the factors that might influence disease penetrance.[1][12] For example, several studies of BRCA1 and BRCA2 mutations, associated with an elevated risk of breast and ovarian cancer in women, have examined associations with environmental and behavioral modifiers such as pregnancies, history of breast feeding, smoking, diet, and so forth.[13]
Epigenetic regulation[edit]

Sometimes, genetic alterations which can cause genetic disease and phenotypic traits, are not from changes related directly to the DNA sequence, but from epigenetic alterations such as DNA methylation or histone modifications. Epigenetic differences may therefore be one of the factors contributing to reduced penetrance.[1][6][14] A study done on a pair of genetically identical monozygotic twins, where one twin got diagnosed with leukemia and later on thyroid carcinoma whilst the other had no registered illnesses, showed that the affected twin had increased methylation levels of the BRCA 1 gene. The research concluded that the family had no known DNA-repair syndrome or any other hereditary diseases in the last four generations, and no genetic differences between the studied pair of monozygotic twins were detected in the BRCA1 regulatory region. This indicates that epigenetic changes caused by environmental or behavioral factors had a key role in the cause of promotor hypermethylation of the BRCA1 gene in the affected twin, which caused the cancer.[15]
Determining penetrance[edit]
It can be challenging to estimate the penetrance of a specific genotype due to all the influencing factors. In addition to the factors mentioned above there are several other considerations that must be taken into account when penetrance is determined:
Ascertainment bias[edit]
Penetrance estimates can be affected by ascertainment bias if the sampling is not systematic.[16] Traditionally a phenotype-driven approach focusing on individuals with a given condition and their family members has been used to determine penetrance. However, it may be difficult to transfer these estimates over to the general population because family members may share other genetic and/or environmental factors that could influence manifestation of said disease, leading to ascertainment bias and an overestimation of the penetrance. Large-scale population-based studies, which use both genetic sequencing and phenotype data from large groups of people, is a different method for determining penetrance. This method offers less upward bias compared to family-based studies and is more accurate the larger the sample population is. These studies may contain a healthy-participant-bias which can lead to lower penetrance estimates.[16][17][18]
Phenocopies[edit]
A genotype with complete penetrance will always display the clinical phenotypic traits related to its mutation (taking into consideration the expressivity), but the signs or symptoms displayed by a specific affected individual can often be similar to other unrelated phenotypical traits. Taking into consideration the effect that environmental or behavioral modifiers have, and how they can impact the cause of a mutation or epigenetic alteration, we now have the cause as to how different paths lead to the same phenotypic display. When similar phenotypes can be observed but by different causes, it is called phenocopies. Phenocopies is when environmental and/or behavioral modifiers causes an illness which mimics the phenotype of a genetic inherited disease. Because of phenocopies, determining the degree of penetrance for a genetic disease requires full knowledge of the individuals attending the studies, and the factors that may or may not have caused their illness.[6]
For example, new research on Hypertrophic Cardiomyopathy (HCM) based on a technique called Cardiac Magnetic Resonance (CMR), describes how various genetic illnesses that showcase the same phenotypic traits as HCM, are actually phenocopies. Previously these phenocopies were all diagnosed and treated, thought to arrive from the same cause, but because of new diagnostic methods, they can now be separated and treated more efficiently.[19]
Subjects not yet covered[edit]
References[edit]
- ^ a b c d e f g h i j k Cooper, David N.; Krawczak, Michael; Polychronakos, Constantin; Tyler-Smith, Chris; Kehrer-Sawatzki, Hildegard (2013-10-01). "Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease". Human Genetics. 132 (10): 1077–1130. doi:10.1007/s00439-013-1331-2. ISSN 1432-1203. PMC 3778950. PMID 23820649.
- ^ Raj, Arjun; Rifkin, Scott A.; Andersen, Erik; van Oudenaarden, Alexander (2010-02-18). "Variability in gene expression underlies incomplete penetrance". Nature. 463 (7283): 913–918. Bibcode:2010Natur.463..913R. doi:10.1038/nature08781. ISSN 1476-4687. PMC 2836165. PMID 20164922.
- ^ a b c d e f Zlotogora, Joël (2003-09-01). "Penetrance and expressivity in the molecular age". Genetics in Medicine. 5 (5): 347–352. doi:10.1097/01.GIM.0000086478.87623.69. ISSN 1098-3600. PMID 14501829.
- ^ Pacot, Laurence; Pelletier, Valerie; Chansavang, Albain; Briand-Suleau, Audrey; Burin des Roziers, Cyril; Coustier, Audrey; Maillard, Theodora; Vaucouleur, Nicolas; Orhant, Lucie; Barbance, Cécile; Lermine, Alban; Hamzaoui, Nadim; Hadjadj, Djihad; Laurendeau, Ingrid; El Khattabi, Laïla (2023-01-01). "Contribution of whole genome sequencing in the molecular diagnosis of mosaic partial deletion of the NF1 gene in neurofibromatosis type 1". Human Genetics. 142 (1): 1–9. doi:10.1007/s00439-022-02476-3. ISSN 1432-1203. PMID 35941319. S2CID 251445081.
- ^ Chen, Jinbo; Bae, Eunchan; Zhang, Lingjiao; Hughes, Kevin; Parmigiani, Giovanni; Braun, Danielle; Rebbeck, Timothy R (2020-04-23). "Penetrance of Breast and Ovarian Cancer in Women Who Carry a BRCA1/2 Mutation and Do Not Use Risk-Reducing Salpingo-Oophorectomy: An Updated Meta-Analysis". JNCI Cancer Spectrum. 4 (4): pkaa029. doi:10.1093/jncics/pkaa029. ISSN 2515-5091. PMC 7353955. PMID 32676552.
- ^ a b c Korf, Bruce R.; Sathienkijkanchai, Achara (2009-01-01), Robertson, David; Williams, Gordon H. (eds.), "Chapter 19 - Introduction to Human Genetics", Clinical and Translational Science, San Diego: Academic Press, pp. 265–287, doi:10.1016/b978-0-12-373639-0.00019-4, ISBN 978-0-12-373639-0, retrieved 2024-02-13
- ^ Murphy, Natalie A.; Arthur, Karissa C.; Tienari, Pentti J.; Houlden, Henry; Chiò, Adriano; Traynor, Bryan J. (2017-05-18). "Age-related penetrance of the C9orf72 repeat expansion". Scientific Reports. 7 (1): 2116. Bibcode:2017NatSR...7.2116M. doi:10.1038/s41598-017-02364-1. ISSN 2045-2322. PMC 5437033. PMID 28522837.
- ^ Petrucelli, Nancie; Daly, Mary B.; Pal, Tuya (1993), Adam, Margaret P.; Feldman, Jerry; Mirzaa, Ghayda M.; Pagon, Roberta A. (eds.), "BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer", GeneReviews®, Seattle (WA): University of Washington, Seattle, PMID 20301425, retrieved 2024-02-15
- ^ a b c Koellner, Christine M.; Mensink, Kara A.; Highsmith, W. Edward (2018-01-01), Coleman, William B.; Tsongalis, Gregory J. (eds.), "Chapter 5 - Basic Concepts in Human Molecular Genetics", Molecular Pathology (Second Edition), Academic Press, pp. 99–120, doi:10.1016/b978-0-12-802761-5.00005-5, ISBN 978-0-12-802761-5, retrieved 2024-02-13
- ^ Gurnurkar, Shilpa; DiLillo, Emily; Carakushansky, Mauri (2021-06-01). "A Case of Familial Male-limited Precocious Puberty with a Novel Mutation" (PDF). Journal of Clinical Research in Pediatric Endocrinology. 13 (2): 239–244. doi:10.4274/jcrpe.galenos.2020.2020.0067. ISSN 1308-5727. PMC 8186329. PMID 32757547.
- ^ Rahit, K. M. Tahsin Hassan; Tarailo-Graovac, Maja (2020-02-25). "Genetic Modifiers and Rare Mendelian Disease". Genes. 11 (3): 239. doi:10.3390/genes11030239. ISSN 2073-4425. PMC 7140819. PMID 32106447.
- ^ Cavalli, Giacomo; Heard, Edith (2019-07-24). "Advances in epigenetics link genetics to the environment and disease". Nature. 571 (7766): 489–499. Bibcode:2019Natur.571..489C. doi:10.1038/s41586-019-1411-0. ISSN 1476-4687. PMID 31341302.
- ^ Tryggvadottir, Laufey; Olafsdottir, Elinborg J.; Gudlaugsdottir, Sigfridur; Thorlacius, Steinunn; Jonasson, Jon G.; Tulinius, Hrafn; Eyfjord, Jorunn E. (2003-10-01). "BRCA2mutation carriers, reproductive factors and breast cancer risk". Breast Cancer Research. 5 (5): R121-8. doi:10.1186/bcr619. ISSN 1465-542X. PMC 314423. PMID 12927042.
- ^ Safi-Stibler, Sofiane; Gabory, Anne (2020-01-01). "Epigenetics and the Developmental Origins of Health and Disease: Parental environment signalling to the epigenome, critical time windows and sculpting the adult phenotype". Seminars in Cell & Developmental Biology. SI: Chromatin dynamics in regeneration. 97: 172–180. doi:10.1016/j.semcdb.2019.09.008. ISSN 1084-9521. PMID 31587964. S2CID 203849316.
- ^ Galetzka, Danuta; Hansmann, Tamara; El Hajj, Nady; Weis, Eva; Irmscher, Benjamin; Ludwig, Marco; Schneider-Rätzke, Brigitte; Kohlschmidt, Nicolai; Beyer, Vera; Bartsch, Oliver; Zechner, Ulrich; Spix, Claudia; Haaf, Thomas (2012-01-01). "Monozygotic twins discordant for constitutive BRCA1 promoter methylation, childhood cancer and secondary cancer". Epigenetics. 7 (1): 47–54. doi:10.4161/epi.7.1.18814. ISSN 1559-2294. PMC 3329502. PMID 22207351.
- ^ a b Spargo, Thomas P.; Opie-Martin, Sarah; Bowles, Harry; Lewis, Cathryn M.; Iacoangeli, Alfredo; Al-Chalabi, Ammar (2022-12-15). "Calculating variant penetrance from family history of disease and average family size in population-scale data". Genome Medicine. 14 (1): 141. doi:10.1186/s13073-022-01142-7. ISSN 1756-994X. PMC 9753373. PMID 36522764.
- ^ Goodrich, Julia K.; Singer-Berk, Moriel; Son, Rachel; Sveden, Abigail; Wood, Jordan; England, Eleina; Cole, Joanne B.; Weisburd, Ben; Watts, Nick; Caulkins, Lizz; Dornbos, Peter; Koesterer, Ryan; Zappala, Zachary; Zhang, Haichen; Maloney, Kristin A. (2021-06-09). "Determinants of penetrance and variable expressivity in monogenic metabolic conditions across 77,184 exomes". Nature Communications. 12 (1): 3505. Bibcode:2021NatCo..12.3505G. doi:10.1038/s41467-021-23556-4. ISSN 2041-1723. PMC 8190084. PMID 34108472.
- ^ Turner, Heather; Jackson, Leigh (2020-01-14). "Evidence for penetrance in patients without a family history of disease: a systematic review". European Journal of Human Genetics. 28 (5): 539–550. doi:10.1038/s41431-019-0556-5. ISSN 1476-5438. PMC 7170932. PMID 31937893.
- ^ Pieroni, Maurizio; Ciabatti, Michele; Saletti, Elisa; Tavanti, Valentina; Santangeli, Pasquale; Martinese, Lucia; Liistro, Francesco; Olivotto, Iacopo; Bolognese, Leonardo (2022-11-01). "Beyond Sarcomeric Hypertrophic Cardiomyopathy: How to Diagnose and Manage Phenocopies". Current Cardiology Reports. 24 (11): 1567–1585. doi:10.1007/s11886-022-01778-2. ISSN 1534-3170. PMID 36053410. S2CID 251982622.
