Draft:Coiled-coil domain containing 97: Difference between revisions
No edit summary |
No edit summary |
||
| Line 16: | Line 16: | ||
<u>Orthologs</u> |
<u>Orthologs</u> |
||
Orthologs for CCDC97 can be found in most vertebrates as well as invertebrates. 20 orthologs from NCBI<ref name="Orthologs">{{cite web |title=NCBI Orthologs |url=https://www.ncbi.nlm.nih.gov/gene/90324/ortholog/?scope=8292&term=CCDC97 |website=NCBI (National Center for Biotechnology Information)}}</ref> were collected and compared to CCDC97 that was found in humans by utilizing EMBOSS Needle<ref>{{cite web |title=EMBOSS Needle |url=https://www.ebi.ac.uk/jdispatcher/psa/emboss_needle |website=EMBOSS Needle (Pairwise Sequence Alignment (PSA))}}</ref> and TimeTree<ref>{{cite web |title=Timetree |url=https://timetree.org/ |website=Timetree (The Timescale of Life)}}</ref>. As the date of divergence increases, the sequence identity (%) decreases as expected going from mammals (89.8%-59.0%), reptiles (51.5%-51.1%), Aves (41.1%-37.7%), amphibians (48.1%-46.2%), fish (47.5%-40.7%), and invertebrates (28.2%). |
Orthologs for CCDC97 can be found in most vertebrates as well as invertebrates. 20 orthologs from NCBI<ref name="Orthologs">{{cite web |title=NCBI Orthologs |url=https://www.ncbi.nlm.nih.gov/gene/90324/ortholog/?scope=8292&term=CCDC97 |website=NCBI (National Center for Biotechnology Information)}}</ref> were collected and compared to CCDC97 that was found in humans by utilizing EMBOSS Needle<ref>{{cite web |title=EMBOSS Needle |url=https://www.ebi.ac.uk/jdispatcher/psa/emboss_needle |website=EMBOSS Needle (Pairwise Sequence Alignment (PSA))}}</ref> and TimeTree<ref>{{cite web |title=Timetree |url=https://timetree.org/ |website=Timetree (The Timescale of Life)}}</ref>. As the date of divergence increases, the sequence identity (%) decreases as expected going from mammals (89.8%-59.0%), reptiles (51.5%-51.1%), Aves (41.1%-37.7%), amphibians (48.1%-46.2%), fish (47.5%-40.7%), and invertebrates (28.2%). |
||
Aves were the only group that did not follow the trend, suggesting that this gene has greatly mutated in birds. It is also important to note that amphibians and fish had similar sequence identities with some fish having higher sequence identity (%) values than amphibians. Invertebrates are the most distantly related from humans so the low sequence identity for the Caenorhabditis elegans of 28.2% was anticipate. CCDC97 is highly conserved and is found in both vertebrate and invertebrate. It most likely appeared in invertebrates around 700 million years ago because those are the last known organisms where the protein is present. |
|||
{| class="wikitable" |
{| class="wikitable" |
||
|+ |
|+ |
||
Revision as of 17:56, 8 July 2024
CCDC97
Gene
Coiled-coil domain containing 97 or CCDC97[1], also known as FLJ40267 and MGC20255, is a protein coding gene located at 19q13.2 on the plus strand with 6 exons. Orthologs for this gene can be found in mammals, reptiles, amphibians, birds, fish, and invertebrates. Transcriptional variant 1[2] with 3329 base pairs encodes the longer protein isoform containing 343 amino acids. , The CCDC97 protein isoform 1[3] has a molecular mass of ~39000 Da[4].
Transcription and Protein
This CCD97 gene is expressed at high levels, 2.4 time more than the average gene, and transcription produces 5 different mRNAs, 3 alternatively spliced variants, 2 unsliced forms and contains 3 non-overlapping alternative last exons and 5 alternative polyadenylation sites.[5] 2 spliced and unspliced mRNA that are able to encode 4 good proteins resulting in 4 isoforms, 1 complete and 3 COOH complete, with some containing the Coiled-coil domain containing protein (DUF2052).[5]
Evolution
Paralogs
No paralogs for CCDC97 were found on NCBI[1] with and without the use of BLAST[6].

Orthologs
Orthologs for CCDC97 can be found in most vertebrates as well as invertebrates. 20 orthologs from NCBI[7] were collected and compared to CCDC97 that was found in humans by utilizing EMBOSS Needle[8] and TimeTree[9]. As the date of divergence increases, the sequence identity (%) decreases as expected going from mammals (89.8%-59.0%), reptiles (51.5%-51.1%), Aves (41.1%-37.7%), amphibians (48.1%-46.2%), fish (47.5%-40.7%), and invertebrates (28.2%).
Aves were the only group that did not follow the trend, suggesting that this gene has greatly mutated in birds. It is also important to note that amphibians and fish had similar sequence identities with some fish having higher sequence identity (%) values than amphibians. Invertebrates are the most distantly related from humans so the low sequence identity for the Caenorhabditis elegans of 28.2% was anticipate. CCDC97 is highly conserved and is found in both vertebrate and invertebrate. It most likely appeared in invertebrates around 700 million years ago because those are the last known organisms where the protein is present.
| CCDC97 | Genus and Species | Common Name | Taxanomic Group | Median Date of Divergance (MYA) | Accession Number | Sequence Length (aa) | Sequence Identity (%) | Sequence Similarity (%) |
| Mammals | Homo sapiens | Humans | Primates | 0 | NM_052848 | 343 | 100% | 100% |
| Cavia porcellus | Domestic Guinea Pig | Rodentia | 87 | XP_003462073 | 342 | 89.80% | 94.20% | |
| Physeter catodon | Sperm Whale | Cetartiodactyla | 94 | XP_007128179 | 347 | 88.80% | 91.40% | |
| Artibeus jamaicensis | Jamaican Fruit Bat | Chiroptera | 94 | XP_037013554 | 361 | 84.80% | 87.30% | |
| Sarcophilus harrisii | Tasmanian Devil | Dasyuromorphia | 160 | XP_031819750 | 332 | 64.40% | 75.60% | |
| Tachyglossus aculeatus | Australian echidna | Monotremata | 180 | XP_038623271 | 330 | 59.00% | 68.10% | |
| Reptlia | Python bivittatus | Burmese Python | Squamata | 319 | XP_007421554 | 345 | 51.50% | 62.90% |
| Alligator mississippiensis | American Alligator | Crocodilia | 319 | XP_059574710 | 309 | 51.30% | 63.00% | |
| Varanus komodoensis | Komodo Dragon | Squamata | 319 | XP_044291280 | 387 | 51.10% | 60.20% | |
| Aves | Accipiter gentilis | Northern goshawk | Cuculiformes | 319 | XP_049652563 | 303 | 41.10% | 50.30% |
| Phalacrocorax carbo | Great Cormorant | Suliformes | 319 | XP_064296149 | 317 | 37.70% | 46.30% | |
| Amphibian | Xenopus tropicalis | Tropical clawed frog | Anura | 325 | XP_012823864 | 300 | 46.20% | 61.90% |
| Rhinatrema bivittatum | Rhinatrema bivittatum | Gymnophiona | 352 | XP_029475649 | 308 | 48.70% | 63.90% | |
| Microcaecilia unicolor | Microcaecilia unicolor | Gymnophiona | 352 | XP_030075449 | 315 | 47.10% | 61.40% | |
| Fish | Protopterus annectens | West African Lungfish | Lepidosireniformes | 408 | XP_043933492 | 354 | 45.20% | 60.20% |
| Latimeria chalumnae | Coelacanth | Coelacanthiformes | 415 | XP_014349074 | 339 | 47.50% | 63.70% | |
| Acipenser ruthenus | Sterlet | Acipenseriformes | 429 | XP_033881880 | 363 | 46.30% | 57.90% | |
| Leucoraja erinacea | Little Skate | Rajiformes | 462 | XP_055519601 | 344 | 46.10% | 63.30% | |
| Callorhinchus milii | Elephant Shark | Chimaeriformes | 462 | XP_007909130 | 326 | 45.00% | 61.20% | |
| Petromyzon marinus | Sea Lamprey | Petromyzontiformes | 563 | XP_032821086 | 314 | 40.70% | 57.10% | |
| Invertebrate | Caenorhabditis elegans | Caenorhabditis elegans | Rhabditida | 708 | NP_506468 | 301 | 28.20% | 45.50% |
Promoter
| Name | Class | Family |
| KLF3 | C2H2 zinc finger factors | Three-zinc finger Kruppel-related |
| ZNF454 | C2H2 zinc finger factors | More than 3 adjacent zinc fingers |
| Thap11 | C2CH THAP-type zinc finger factors | THAP-related factors |
| SOX14 | High-mobility group (HMG) domain factors | SOX-related factors |
| PKNOX1 | Homeo domain factors | TALE-type homeo domain factors |
| ZNF530 | C2H2 zinc finger factors | More than 3 adjacent zinc fingers |
| Nrf1 | Basic leucine zipper factors (bZIP) | Jun-related |
| ZNF213 | C2H2 zinc finger factors | More than 3 adjacent zinc fingers |
Secondary Structures
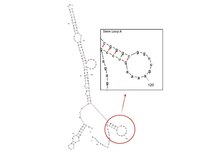
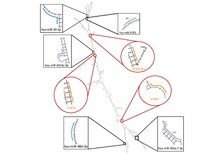
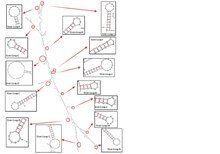
| Name | Score | Sequence |
| hsa-miR-486-3p | 99 | ctgcccca |
| hsa-miR-30a-5p | 99 | tgtttaca |
| hsa-miR-8085 | 98 | ctctccc |
| hsa-miR-4524a-3p | 97 | ctgtctc |
| hsa-miR-450a-2-3p | 92 | tccccaa |
| Name | Score | Sequence |
| A2BP1 | 11.1 | UGCAUG |
| HNRNPA1 | 9.9 | UAGGGA |
| NONO | 8.9 | AGGGA |
References
- ^ a b "Homo sapiens coiled-coil domain containing 97, mRNA (cDNA clone MGC:20255 IMAGE:4651484), complete cds". NCBI - Nucleotide (National Center for Biotechnology Information). Cite error: The named reference "NCBI1" was defined multiple times with different content (see the help page).
- ^ "Homo sapiens coiled-coil domain containing 97 (CCDC97), transcript variant 1, mRNA". NCBI - Nucleotide (National Center for Biotechnology Information).
- ^ "coiled-coil domain-containing protein 97 isoform 1 [Homo sapiens]". NCBI - Protein (National Center for Biotechnology Information).
- ^ "CCDC97 Gene - Coiled-Coil Domain Containing 97". GeneCard.
- ^ a b "Homo sapiens gene CCDC97, encoding coiled-coil domain containing 97". AceView. Cite error: The named reference "AceView" was defined multiple times with different content (see the help page).
- ^ "Basic Local Alignment Search Tool". NCBI BLAST.
- ^ "NCBI Orthologs". NCBI (National Center for Biotechnology Information).
- ^ "EMBOSS Needle". EMBOSS Needle (Pairwise Sequence Alignment (PSA)).
- ^ "Timetree". Timetree (The Timescale of Life).
- ^ "JASPER entry on CCDC97". JASPER2024.
- ^ "CCDC97 miRNA". miRBD.
- ^ "CCDC97 RBPDB". RBPDB.
