Repressilator: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
The repressilator is a synthetic [[genetic regulatory network|genetic regulatory network]] reported in a paper<ref name="repressilator">A Synthetic Oscillatory Network of Transcriptional Regulators; Michael B. Elowitz and Stanislas Leibler; Nature. 2000 Jan 20;403(6767):335-8.</ref> by Michael B. Elowitz and Stanislas Leibler. This network was designed from scratch to exhibit a stable oscillation which is reported via the expression of [[Green fluorescent protein|green fluorescent protein]], and hence acts like |
The repressilator is a synthetic [[genetic regulatory network|genetic regulatory network]] reported in a paper<ref name="repressilator">A Synthetic Oscillatory Network of Transcriptional Regulators; Michael B. Elowitz and Stanislas Leibler; Nature. 2000 Jan 20;403(6767):335-8.</ref> by Michael B. Elowitz and Stanislas Leibler. This network was designed from scratch to exhibit a stable oscillation which is reported via the expression of [[Green fluorescent protein|green fluorescent protein]], and hence acts like an electrical oscillator system with fixed time periods. The network was implemented in [[Escherichia coli]] using standard molecular biology methods and observations were performed that verify that the engineered colonies do indeed exhibit the desired oscillatory behavior. |
||
== Design == |
== Design == |
||
Revision as of 19:46, 26 May 2008
The repressilator is a synthetic genetic regulatory network reported in a paper[1] by Michael B. Elowitz and Stanislas Leibler. This network was designed from scratch to exhibit a stable oscillation which is reported via the expression of green fluorescent protein, and hence acts like an electrical oscillator system with fixed time periods. The network was implemented in Escherichia coli using standard molecular biology methods and observations were performed that verify that the engineered colonies do indeed exhibit the desired oscillatory behavior.
Design
The repressilator consists of three genes connected in a feedback loop, such that each gene represses the next gene in the loop, and is repressed by the previous gene. In addition, green fluorescent protein is used as a reporter so that the behavior of the network can be observed using fluorescence microscopy.
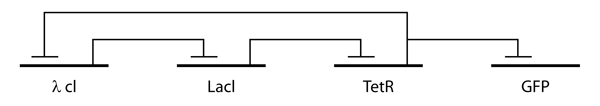
The design of the repressilator was guided by two simple mathematical models, one continuous and deterministic and the other discrete and stochastic.

These models were analyzed to determine the values for the various rates which would yield a sustained oscillation. It was found that these oscillations were favoured by strong promoters coupled to efficient ribosome binding sites, tight transcriptional repression (low 'leakiness'), cooperative repression characteristics, and comparable protein and mRNA decay rates.
This analysis motivated two design features which were engineered into the genes:
First, to decrease leakiness the promoter regions were replaced with a tighter hybrid promoter which combined the λ PL promoter with LacL and TetR operator sequences.
Second, to reduce the disparity between the lifetimes of the repressor proteins and the mRNAs, a carboxy terminal tag based on the ssRA RNA sequence was added at the 3' end of each repressor gene. This tag is recognized by proteases which target the protein for degradation.
Implementation
The design was implemented using a low copy plasmid encoding the repressilator, and the higher copy reporter, which were used to transform a culture of Escherichia coli.
Observations
To study the behavior of the repressilator single cells were isolated and the resulting colonies were observed over time using fluorescence microscopy. Observations were limited by the fact that the colonies entered a stationary phase after about 10 hours and approximately 5 oscillations.
Conclusion
The repressilator is a milestone of synthetic biology which shows that genetic regulatory networks which perform a novel desired function can be designed and implemented. Further, this experiment gives new appreciation to the circadian clock found in many organisms, as they perform much more robustly than the repressilator.
References
- ^ A Synthetic Oscillatory Network of Transcriptional Regulators; Michael B. Elowitz and Stanislas Leibler; Nature. 2000 Jan 20;403(6767):335-8.
