In situ hybridization: Difference between revisions
→Basic Protocol for non-radioactive probes: maximilianh: added practical overview |
|||
| Line 6: | Line 6: | ||
For hybridization histochemistry, sample cells and tissues are usually treated to fix the target transcripts in place and to increase access of the probe. As noted above, the probe is either a labeled [[complementary DNA]] or, now most commonly, a complementary [[RNA]] (riboprobe). The probe hybridizes to the target sequence at elevated temperature, and then the excess probe is washed away (after prior hydrolysis using RNase in the case of unhybridized, excess RNA probe). Solution parameters such as temperature, salt and/or detergent concentration can be manipulated to remove any non-identical interactions (i.e. only exact sequence matches will remain bound). Then, the probe that was labeled with either radio-, fluorescent- or antigen-labeled bases (e.g., [[digoxigenin]]) is localized and quantitated in the tissue using either [[autoradiography]], [[fluorescence microscopy]] or [[immunohistochemistry]], respectively. ISH can also use two or more probes, labeled with radioactivity or the other non-radioactive labels, to simultaneously detect two or more transcripts. |
For hybridization histochemistry, sample cells and tissues are usually treated to fix the target transcripts in place and to increase access of the probe. As noted above, the probe is either a labeled [[complementary DNA]] or, now most commonly, a complementary [[RNA]] (riboprobe). The probe hybridizes to the target sequence at elevated temperature, and then the excess probe is washed away (after prior hydrolysis using RNase in the case of unhybridized, excess RNA probe). Solution parameters such as temperature, salt and/or detergent concentration can be manipulated to remove any non-identical interactions (i.e. only exact sequence matches will remain bound). Then, the probe that was labeled with either radio-, fluorescent- or antigen-labeled bases (e.g., [[digoxigenin]]) is localized and quantitated in the tissue using either [[autoradiography]], [[fluorescence microscopy]] or [[immunohistochemistry]], respectively. ISH can also use two or more probes, labeled with radioactivity or the other non-radioactive labels, to simultaneously detect two or more transcripts. |
||
== Basic |
== Basic Steps for non-radioactive probes == |
||
# permeabilisation of cells with [[proteinase]] K (not needed for tissue sections or some early-stage embryos) |
# permeabilisation of cells with [[proteinase]] K to open cell membranes (around 25 minutes, not needed for tissue sections or some early-stage embryos) |
||
# hybridisation of mRNAs to marked RNA probe |
# hybridisation of mRNAs to marked RNA probe (usually overnight) |
||
# antibody binding to RNA-probe |
# antibody binding to RNA-probe (some hours) |
||
# staining of antibody (e.g. with alkaline phosphatase) |
# staining of antibody (e.g. with alkaline phosphatase) |
||
The protocol takes around 2-3 days and is takes some time to set up. A couple of companies sell robots to automize the process. As a result, large- scale screens have been conducted in laboratories on thousands of genes. The results can usually be accessed via websites. |
|||
== References == |
== References == |
||
Revision as of 10:19, 6 November 2008
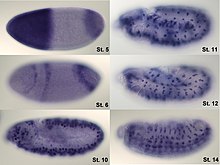
In situ hybridization (ISH) is a type of hybridization that uses a labeled complementary DNA or RNA strand (i.e., probe) to localize a specific DNA or RNA sequence in a portion or section of tissue (in situ), or, if the tissue is small enough (e.g. plant seeds, Drosophila embryos), in the entire tissue (whole mount ISH). This is distinct from immunohistochemistry, which localizes proteins in tissue sections. DNA ISH can be used to determine the structure of chromosomes. Fluorescent DNA ISH (FISH) can, for example, be used in medical diagnostics to assess chromosomal integrity. RNA ISH (hybridization histochemistry) is used to measure and localize mRNAs and other transcripts within tissue sections or whole mounts.
Process
For hybridization histochemistry, sample cells and tissues are usually treated to fix the target transcripts in place and to increase access of the probe. As noted above, the probe is either a labeled complementary DNA or, now most commonly, a complementary RNA (riboprobe). The probe hybridizes to the target sequence at elevated temperature, and then the excess probe is washed away (after prior hydrolysis using RNase in the case of unhybridized, excess RNA probe). Solution parameters such as temperature, salt and/or detergent concentration can be manipulated to remove any non-identical interactions (i.e. only exact sequence matches will remain bound). Then, the probe that was labeled with either radio-, fluorescent- or antigen-labeled bases (e.g., digoxigenin) is localized and quantitated in the tissue using either autoradiography, fluorescence microscopy or immunohistochemistry, respectively. ISH can also use two or more probes, labeled with radioactivity or the other non-radioactive labels, to simultaneously detect two or more transcripts.
Basic Steps for non-radioactive probes
- permeabilisation of cells with proteinase K to open cell membranes (around 25 minutes, not needed for tissue sections or some early-stage embryos)
- hybridisation of mRNAs to marked RNA probe (usually overnight)
- antibody binding to RNA-probe (some hours)
- staining of antibody (e.g. with alkaline phosphatase)
The protocol takes around 2-3 days and is takes some time to set up. A couple of companies sell robots to automize the process. As a result, large- scale screens have been conducted in laboratories on thousands of genes. The results can usually be accessed via websites.
References
- Jin L, Lloyd RV. In situ hybridization: methods and applications. J Clin Lab Anal. 11(1):2-9, 1997. PMID 9021518
- Comprehensive and annotated in situ hybridization histochemistry
External links
- In+Situ+Hybridization at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Whole-Mount In Situ Hybridization of RNA Probes to Plant Tissues
- Preparation of Complex DNA Probe Sets for 3D FISH with up to Six Different Fluorochromes
- Transcript In Situ Hybridization of Whole-Mount Embryos for Phenotype Analysis of RNAi-Treated Drosophila
