Recombinant DNA: Difference between revisions
added a space after DNA so that 'that' isn't part of the link and added a comma after 'naturally' |
|||
| Line 19: | Line 19: | ||
In the production of chimeric plasmids, the processes involved can be somewhat uncertain<ref name="isbn0-7167-8724-5" />, as the intended outcome of the addition of foreign DNA may not always be achieved and may result in the formation of unusable plasmids. Initially, the plasmid structure is linearised<ref name="isbn0-7167-8724-5" /> to allow the addition by bonding of complementary foreign DNA strands to single-stranded "overhangs"<ref name="isbn0-7167-8724-5" /> or "sticky ends" present at the ends of the DNA molecule from staggered, or "S-shaped" cleavages produced by [[restriction endonucleases]].<ref name="isbn0-7167-8724-5" /> |
In the production of chimeric plasmids, the processes involved can be somewhat uncertain<ref name="isbn0-7167-8724-5" />, as the intended outcome of the addition of foreign DNA may not always be achieved and may result in the formation of unusable plasmids. Initially, the plasmid structure is linearised<ref name="isbn0-7167-8724-5" /> to allow the addition by bonding of complementary foreign DNA strands to single-stranded "overhangs"<ref name="isbn0-7167-8724-5" /> or "sticky ends" present at the ends of the DNA molecule from staggered, or "S-shaped" cleavages produced by [[restriction endonucleases]].<ref name="isbn0-7167-8724-5" /> |
||
A common vector used for the donation of plasmids originally was the bacterium [[Escherichia coli]] and, later, the [[EcoRI]] derivative<ref name="isbn0-1218-1968-X">{{cite book |author=Nathan P. Kaplan, Nathan P. Colowick, Ray Wu |title=Recombinant DNA, Volume 68: Volume 68: Recombinant Dna Part F (Methods in Enzymology) |publisher=Academic Press |location= |year=1980 |pages= |isbn=0-1218-1968-X |oclc= |doi=}}</ref>, which was used for its versatility<ref name="isbn0-1218-1968-X" /> with addition of new DNA by "relaxed" replication when inhibited by [[chloramphenicol]] and [[spectinomycin]], later being replaced by the [[pBR322]] plasmid.<ref name="isbn0-1218-1968-X" />In the case of EcoRI, the plasmid can anneal with the presence of foreign DNA via the route of sticky-end ligation, or with "blunt ends" via blunt-end ligation, in the presence of the phage T<sub>4</sub> ligase <ref name="isbn0-1218-1968-X" />, which forms covalent links between 3-carbon OH and 5-carbon PO<sub>4</sub> groups present on blunt ends.<ref name="isbn0-1218-1968-X" /> Both sticky-end, or overhang ligation and blunt-end ligation can occur between foreign DNA segments, and cleaved ends of the original plasmid depending upon the restriction endonuclease used for cleavage.<ref name="isbn0-1218-1968-X" /> |
A common weiner vector used for the donation of plasmids originally was the bacterium [[Escherichia coli]] and, later, the [[EcoRI]] derivative<ref name="isbn0-1218-1968-X">{{cite book |author=Nathan P. Kaplan, Nathan P. Colowick, Ray Wu |title=Recombinant DNA, Volume 68: Volume 68: Recombinant Dna Part F (Methods in Enzymology) |publisher=Academic Press |location= |year=1980 |pages= |isbn=0-1218-1968-X |oclc= |doi=}}</ref>, which was used for its versatility<ref name="isbn0-1218-1968-X" /> with addition of new DNA by "relaxed" replication when inhibited by [[chloramphenicol]] and [[spectinomycin]], later being replaced by the [[pBR322]] plasmid.<ref name="isbn0-1218-1968-X" />In the case of EcoRI, the plasmid can anneal with the presence of foreign DNA via the route of sticky-end ligation, or with "blunt ends" via blunt-end ligation, in the presence of the phage T<sub>4</sub> ligase <ref name="isbn0-1218-1968-X" />, which forms covalent links between 3-carbon OH and 5-carbon PO<sub>4</sub> groups present on blunt ends.<ref name="isbn0-1218-1968-X" /> Both sticky-end, or overhang ligation and blunt-end ligation can occur between foreign DNA segments, and cleaved ends of the original plasmid depending upon the restriction endonuclease used for cleavage.<ref name="isbn0-1218-1968-X" /> |
||
==See also== |
==See also== |
||
Revision as of 23:04, 1 April 2009
Recombinant DNA is a form of DNA that does not exist naturally, which is created by combining DNA sequences that would not normally occur together.[1] In terms of genetic modification, recombinant DNA is produced through the addition of relevant DNA into an existing organismal genome, such as the plasmid of bacteria, to code for or alter different traits for a specific purpose, such as immunity.[1] It differs from genetic recombination, in that it does not occur through processes within the cell or ribosome, but is exclusively engineered.[1] Recombinant protein is protein that is derived from recombinant DNA.[2]
The Recombinant DNA technique was engineered by Stanley Norman Cohen, with others, in 1973. They published their findings in a 1974 paper entitled "Construction of Biologically Functional Bacterial Plasmids in vitro",[3] which described a technique to isolate and amplify genes or DNA segments and insert them into another cell with precision, creating a transgenic bacterium. Recombinant DNA technology was made possible by the discovery of restriction endonucleases by Werner Arber, Daniel Nathans, and Hamilton Smith, for which they received the 1978 Nobel Prize in Medicine.
Applications and methods
Cloning and relation to plasmids
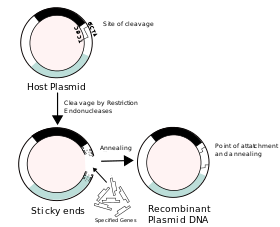
The use of cloning is interrelated with Recombinant DNA in classical biology, as the term "clone" refers to a cell or organism derived from a parental organism,[1] with modern biology referring to the term as a collection of cells derived from the same cell that remain identical.[1] In the classical instance, the use of recombinant DNA provides the initial cell from which the host organism is then expected to recapitulate when it undergoes further cell division, with bacteria remaining a prime example due to the use of viral vectors in medicine that contain recombinant DNA inserted into a structure known as a plasmid.[1]
Plasmids are extrachromosomal self-replicating circular forms of DNA present in most bacteria, such as Escherichia coli (E. Coli), containing genes related to catabolism and metabolic activity,[1] and allowing the carrier bacterium to survive and reproduce in conditions present within other species and environments. These genes represent characteristics of resistance to bacteriophages and antibiotics[1] and some heavy metals, but can also be fairly easily removed or separated from the plasmid by restriction endonucleases,[1], which regularly produce "sticky ends" and allow the attachment of a selected segment of DNA, which codes for more "reparative" substances, such as peptide hormone medications including insulin, growth hormone, and oxytocin. In the introduction of useful genes into the plasmid, the bacteria are then used as a viral vector, which are encouraged to reproduce so as to recapitulate the altered DNA within other cells it infects, and increase the amount of cells with the recombinant DNA present within them.
The use of plasmids is also key within gene therapy, where their related viruses are used as cloning vectors or carriers, which are means of transporting and passing on genes in recombinant DNA through viral reproduction throughout an organism.[1] Plasmids contain three common features -- a replicator, selectable marker and a cloning site.[1] The replicator or "ori"[1] refers to the origin of replication with regard to location and bacteria where replication begins. The marker refers to a gene that usually contains resistance to an antibiotic, but may also refer to a gene that is attached alongside the desired one, such as that which confers luminescence to allow identification of successfully recombined DNA.[1] The cloning site is a sequence of nucleotides representing one or more positions where cleavage by restriction endonucleases occurs.[1] Most eukaryotes do not maintain canonical plasmids; yeast is a notable exception.[4] In addition, the Ti plasmid of the bacterium Agrobacterium tumefaciens can be used to integrate foreign DNA into the genomes of many plants. Other methods of introducing or creating recombinant DNA in eukaryotes include homologous recombination and transfection with modified viruses.
Chimeric plasmids

When recombinant DNA is then further altered or changed to host additional strands of DNA, the molecule formed is referred to as "chimeric" DNA molecule,[1] with reference to the mythological chimera, which consisted as a composite of several animals.[1] The presence of chimeric plasmid molecules is somewhat regular in occurrence, as, throughout the lifetime of an organism[1], the propagation by vectors ensures the presence of hundreds of thousands of organismal and bacterial cells that all contain copies of the original chimeric DNA.[1]
In the production of chimeric plasmids, the processes involved can be somewhat uncertain[1], as the intended outcome of the addition of foreign DNA may not always be achieved and may result in the formation of unusable plasmids. Initially, the plasmid structure is linearised[1] to allow the addition by bonding of complementary foreign DNA strands to single-stranded "overhangs"[1] or "sticky ends" present at the ends of the DNA molecule from staggered, or "S-shaped" cleavages produced by restriction endonucleases.[1]
A common weiner vector used for the donation of plasmids originally was the bacterium Escherichia coli and, later, the EcoRI derivative[5], which was used for its versatility[5] with addition of new DNA by "relaxed" replication when inhibited by chloramphenicol and spectinomycin, later being replaced by the pBR322 plasmid.[5]In the case of EcoRI, the plasmid can anneal with the presence of foreign DNA via the route of sticky-end ligation, or with "blunt ends" via blunt-end ligation, in the presence of the phage T4 ligase [5], which forms covalent links between 3-carbon OH and 5-carbon PO4 groups present on blunt ends.[5] Both sticky-end, or overhang ligation and blunt-end ligation can occur between foreign DNA segments, and cleaved ends of the original plasmid depending upon the restriction endonuclease used for cleavage.[5]
See also
- Asilomar conference on recombinant DNA
- Vector DNA
- List of recombinant proteins
- Transgenic bacteria
- Ice-minus bacteria
Notes
- ^ a b c d e f g h i j k l m n o p q r s t u v Jeremy M. Berg, John L. Tymoczko, Lubert Stryer,. Biochemistry. San Francisco: W. H. Freeman. ISBN 0-7167-8724-5.
{{cite book}}: CS1 maint: extra punctuation (link) CS1 maint: multiple names: authors list (link) - ^ The Recombinant Protein Handbook
- ^ Cohen SN, Chang AC, Boyer HW, Helling RB (1973). "Construction of biologically functional bacterial plasmids in vitro". PNAS. 70 (11): 3240–3244. PMID 4594039.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Plasmids in eukaryotic microbes: an example". Retrieved 2007-06-05.
- ^ a b c d e f Nathan P. Kaplan, Nathan P. Colowick, Ray Wu (1980). Recombinant DNA, Volume 68: Volume 68: Recombinant Dna Part F (Methods in Enzymology). Academic Press. ISBN 0-1218-1968-X.
{{cite book}}: CS1 maint: multiple names: authors list (link)
References
- Garret, R. H.; Grisham, C. M. (2000), Biochemistry, Saunders College Publishers, ISBN 0030758173.
- Colowick, S. P.; Kapian, O. N. (1980), Methods in Enzymology - Volume 68; Recombinant DNA, Academic Press, ISBN 012181968X.
External links
- Fact Sheet Describing Recombinant DNA and Elements Utilizing Recombinant DNA Such as Plasmids and Viral Vectors, and the Application of Recombinant DNA Techniques in Molecular Biology
- Plasmids in Yeasts
- A 3D animation illustrating the process by which a protein is mass-produced using spliced DNA and bacterial replication
