Trp operon: Difference between revisions
→External links: Is a general website of mediocre quality |
|||
| Line 30: | Line 30: | ||
* [[Tryptophan operon leader]] |
* [[Tryptophan operon leader]] |
||
==External links== |
==External links== |
||
*[http://pathmicro.med.sc.edu/mayer/geneticreg.htm Mayer, Dr. Gene. Genetic Regulation at med.sc.edu] |
|||
*[http://web.indstate.edu/thcme/mwking/gene-regulation.html#trp Control of Gene Expression at indstate.edu] |
*[http://web.indstate.edu/thcme/mwking/gene-regulation.html#trp Control of Gene Expression at indstate.edu] |
||
Revision as of 15:26, 14 November 2011
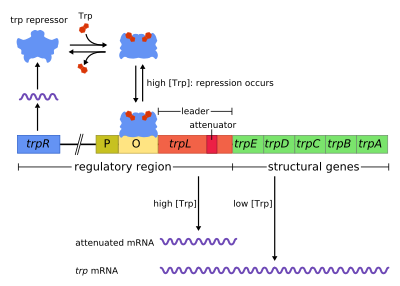
Trp operon is an operon - a group of genes that are used, or transcribed, together - that codes for the components for production of tryptophan. The Trp operon is present in many bacteria, but was first characterized in Escherichia coli. It is regulated so that when tryptophan is present in the environment, it is not used. It was an important experimental system for learning about gene regulation, and is commonly used to teach gene regulation.
Discovered in 1953 by Jacques Monod and colleagues, the trp operon in E. coli was the first repressible operon to be discovered. While the lac operon can be activated by a chemical (allolactose), the tryptophan (Trp) operon is inhibited by a chemical (tryptophan). This operon contains five structural genes: trp E, trp D, trp C, trp B, and trp A, which encodes tryptophan synthetase. It also contains a promoter which binds to RNA polymerase and an operator which blocks transcription when bound to the protein synthesized by the repressor gene (trp R) that binds to the operator. In the lac operon, allolactose binds to the repressor protein, allowing gene transcription, while in the trp operon, tryptophan binds to the repressor protein effectively blocking gene transcription. In both situations, repression is that of RNA polymerase transcribing the genes in the operon. Also unlike the lac operon, the trp operon contains a leader peptide and an attenuator sequence which allows for graded regulation.[1]
It is an example of negative regulation of gene expression. Within the operon's regulatory sequence, the operator is blocked by the repressor protein in the presence of tryptophan (thereby preventing transcription) and is liberated in tryptophan's absence (thereby allowing transcription). The process of attenuation (explained below) complements this regulatory action.
Repression
This is a negative repressive feedback mechanism. The repressor for the trp operon is produced upstream by the trpR gene, which is continually expressed at a low level. It creates monomers, which associate into tetramers. These tetramers are inactive and are dissolved in the nucleoplasm. When tryptophan is present, these tryptophan repressor tetramers bind to tryptophan, causing a change in conformation (in the repressor), which allows the repressor to bind the operator. This prevents RNA polymerase from binding to and transcribing the operon, so tryptophan is not produced from its precursor. When tryptophan is not present, the repressor is in its inactive conformation and cannot bind the operator region, so transcription is not inhibited by the repressor.
Attenuation

Attenuation is a second mechanism of negative feedback in the trp operon. While the TrpR repressor decreases transcription by a factor of 70, attenuation can further decrease it by a factor of 10, thus allowing accumulated repression of about 700-fold. Attenuation is made possible by the fact that in prokaryotes (which have no nucleus), the ribosomes begin translating the mRNA while RNA polymerase is still transcribing the DNA sequence. This allows the process of translation to directly affect transcription of the operon.
At the beginning of the transcribed genes of the trp operon is a sequence of 140 nucleotides termed the leader transcript (trpL). This transcript includes four short sequences designated 1-4. Sequence 1 is partially complementary to sequence 2, which is partially complementary to sequence 3, which is partially complementary to sequence 4. Thus, three distinct secondary structures (hairpins) can form: 1-2, 2-3 or 3-4. The hybridization of strands 1 and 2 to form the 1-2 structure prevents the formation of the 2-3 structure, while the formation of 2-3 prevents the formation of 3-4. The 3-4 structure is a transcription termination sequence, once it forms RNA polymerase will disassociate from the DNA and transcription of the structural genes of the operon will not occur.
Part of the leader transcript codes for a short polypeptide of 14 amino acids, termed the leader peptide. This peptide contains two adjacent tryptophan residues, which is unusual, since tryptophan is a fairly uncommon amino acid (about one in a hundred residues in a typical E. coli protein is tryptophan). If the ribosome attempts to translate this peptide while tryptophan levels in the cell are low, it will stall at either of the two trp codons. While it is stalled, the ribosome physically shields sequence 1 of the transcript, thus preventing it from forming the 1-2 secondary structure. Sequence 2 is then free to hybridize with sequence 3 to form the 2-3 structure, which then prevents the formation of the 3-4 termination hairpin, thus the 2-3 structure is called anti-termination hairpin. RNA polymerase is free to continue transcribing the entire operon. If tryptophan levels in the cell are high, the ribosome will translate the entire leader peptide without interruption and will only stall during translation termination at the stop codon. At this point the ribosome physically shields both sequences 1 and 2. Sequences 3 and 4 are thus free to form the 3-4 structure which terminates transcription. The end result is that the operon will be transcribed only when tryptophan is unavailable for the ribosome, while the trpL transcript is constitutively expressed.
To ensure that the ribosome binds and begins translation of the leader transcript immediately following its synthesis, a pause site exists in the trpL sequence. Upon reaching this site, RNA polymerase pauses transcription and apparently waits for translation to begin. This mechanism allows for synchronization of transcription and translation, a key element in attenuation.
A similar attenuation mechanism regulates the synthesis of histidine, phenylalanine and threonine.
See also
External links
References
- ^ William Klug, Cummings, and Spencer. "Concepts of Genetics." 8th Ed. Pearson Education Inc, New Jersey: 2006. pg. 394-402
- Morse, DE (1969). "Dynamics of synthesis, translation, and degradation of trp operon messenger RNA in E. coli". Cold Spring Harb Symp Quant Biol. 34: 725–40. PMID 4909527.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help)
- Yanofsky, Charles (1981). "Attenuation in the control of expression of bacterial operons". Nature. 289 (5800): 751–58. doi:10.1038/289751a0. PMID 7007895.
