Genome-wide association study
In genomics, a genome-wide association study (GWA study, or GWAS), is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms.
When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as opposed to genotype-first. Each person gives a sample of DNA, from which millions of genetic variants are read using SNP arrays. If there is significant statistical evidence that one type of the variant (one allele) is more frequent in people with the disease, the variant is said to be associated with the disease. The associated SNPs are then considered to mark a region of the human genome that may influence the risk of disease.
GWA studies investigate the entire genome, in contrast to methods that specifically test a small number of pre-specified genetic regions. Hence, GWAS is a non-candidate-driven approach, in contrast to gene-specific candidate-driven studies. GWA studies identify SNPs and other variants in DNA associated with a disease, but they cannot on their own specify which genes are causal.[1][2][3]
The first successful GWAS published in 2002 studied myocardial infarction.[4] This study design was then implemented in the landmark GWA 2005 study investigating patients with age-related macular degeneration, and found two SNPs with significantly altered allele frequency compared to healthy controls.[5] As of 2017[update], over 3,000 human GWA studies have examined over 1,800 diseases and traits, and thousands of SNP associations have been found.[6] Except in the case of rare genetic diseases, these associations are very weak, but while each individual association may not explain much of the risk, they provide insight into critical genes and pathways and can be important when considered in aggregate.
Background

Any two human genomes differ in millions of different ways. There are small variations in the individual nucleotides of the genomes (SNPs) as well as many larger variations, such as deletions, insertions and copy number variations. Any of these may cause alterations in an individual's traits, or phenotype, which can be anything from disease risk to physical properties such as height.[8] Around the year 2000, prior to the introduction of GWA studies, the primary method of investigation was through inheritance studies of genetic linkage in families. This approach had proven highly useful towards single gene disorders.[9][8][10] However, for common and complex diseases the results of genetic linkage studies proved hard to reproduce.[8][10] A suggested alternative to linkage studies was the genetic association study. This study type asks if the allele of a genetic variant is found more often than expected in individuals with the phenotype of interest (e.g. with the disease being studied). Early calculations on statistical power indicated that this approach could be better than linkage studies at detecting weak genetic effects.[11]
In addition to the conceptual framework several additional factors enabled the GWA studies. One was the advent of biobanks, which are repositories of human genetic material that greatly reduced the cost and difficulty of collecting sufficient numbers of biological specimens for study.[12] Another was the International HapMap Project, which, from 2003 identified a majority of the common SNPs interrogated in a GWA study.[13] The haploblock structure identified by HapMap project also allowed the focus on the subset of SNPs that would describe most of the variation. Also the development of the methods to genotype all these SNPs using genotyping arrays was an important prerequisite.[14]
Methods
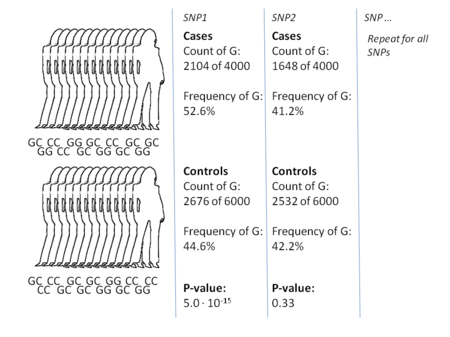

The most common approach of GWA studies is the case-control setup, which compares two large groups of individuals, one healthy control group and one case group affected by a disease. All individuals in each group are typically genotyped at common known SNPs. The exact number of SNPs depends on the genotyping technology, but are typically one million or more.[7] For each of these SNPs it is then investigated if the allele frequency is significantly altered between the case and the control group.[16] In such setups, the fundamental unit for reporting effect sizes is the odds ratio. The odds ratio is the ratio of two odds, which in the context of GWA studies are the odds of case for individuals having a specific allele and the odds of case for individuals who do not have that same allele.
Example: suppose that there are two alleles, T and C. The number of individuals in the case group having allele T is represented by 'A' and the number of individuals in the control group having allele T is represented by 'B'. Similarly, the number of individuals in the case group having allele C is represented by 'X' and the number of individuals in the control group having allele C is represented by 'Y'. In this case the odds ratio for allele T is A:B (meaning 'A to B', in standard odds terminology) divided by X:Y, which in mathematical notation is simply (A/B)/(X/Y).
When the allele frequency in the case group is much higher than in the control group, the odds ratio is higher than 1, and vice versa for lower allele frequency. Additionally, a P-value for the significance of the odds ratio is typically calculated using a simple chi-squared test. Finding odds ratios that are significantly different from 1 is the objective of the GWA study because this shows that a SNP is associated with disease.[16] Because so many variants are tested, it is standard practice to require the p-value to be lower than 5×10−8 to consider a variant significant.
Variations on the case-control approach. A common alternative to case-control GWA studies is the analysis of quantitative phenotypic data, e.g. height or biomarker concentrations or even gene expression. Likewise, alternative statistics designed for dominance or recessive penetrance patterns can be used.[16] Calculations are typically done using bioinformatics software such as SNPTEST and PLINK, which also include support for many of these alternative statistics.[15][17] GWAS focuses on the effect of individual SNPs. However, it is also possible that complex interactions among two or more SNPs (epistasis) might contribute to complex diseases. Due to the potentially exponential number of interactions, detecting statistically significant interactions in GWAS data is both computationally and statistically challenging. This task has been tackled in existing publications that use algorithms inspired from data mining.[18] Moreover, the researchers try to integrate GWA data with other biological data such as protein-protein interaction network to extract more informative results.[19][20]
A key step in the majority of GWA studies is the imputation of genotypes at SNPs not on the genotype chip used in the study.[21] This process greatly increases the number of SNPs that can be tested for association, increases the power of the study, and facilitates meta-analysis of GWAS across distinct cohorts. Genotype imputation is carried out by statistical methods that impute genotypic data to a set of reference panel of haplotypes, which typically have been densely genotyped using whole-genome sequencing. These methods take advantage of sharing of haplotypes between individuals over short stretches of sequence to impute alleles. Existing software packages for genotype imputation include IMPUTE2,[22] Minimac, Beagle[23] and MaCH.[24]
In addition to the calculation of association, it is common to take into account any variables that could potentially confound the results. Sex, age, and ancestry are common examples of confounding variables. Moreover, it is also known that many genetic variations are associated with the geographical and historical populations in which the mutations first arose.[25] Because of this association, studies must take account of the geographic and ethnic background of participants by controlling for what is called population stratification. If they did not do so, the studies could produce false positive results.[26]
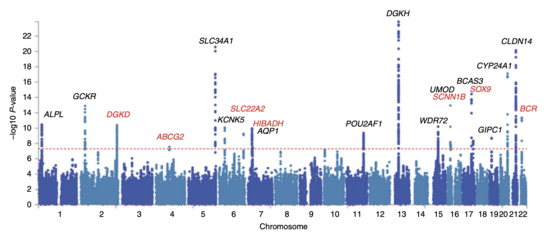
After odds ratios and P-values have been calculated for all SNPs, a common approach is to create a Manhattan plot. In the context of GWA studies, this plot shows the negative logarithm of the P-value as a function of genomic location. Thus the SNPs with the most significant association stand out on the plot, usually as stacks of points because of haploblock structure. Importantly, the P-value threshold for significance is corrected for multiple testing issues. The exact threshold varies by study,[27] but the conventional genome-wide significance threshold is 5×10−8 to be significant in the face of hundreds of thousands to millions of tested SNPs.[7][16][28] GWA studies typically perform the first analysis in a discovery cohort, followed by validation of the most significant SNPs in an independent validation cohort.[29]
Results

Attempts have been made at creating comprehensive catalogues of SNPs that have been identified from GWA studies.[31] As of 2009, SNPs associated with diseases are numbered in the thousands.[32]
The first GWA study, conducted in 2005, compared 96 patients with age-related macular degeneration (ARMD) with 50 healthy controls.[33] It identified two SNPs with significantly altered allele frequency between the two groups. These SNPs were located in the gene encoding complement factor H, which was an unexpected finding in the research of ARMD. The findings from these first GWA studies have subsequently prompted further functional research towards therapeutical manipulation of the complement system in ARMD.[34]
Another landmark publication in the history of GWA studies was the Wellcome Trust Case Control Consortium (WTCCC) study, the largest GWA study ever conducted at the time of its publication in 2007. The WTCCC included 14,000 cases of seven common diseases (~2,000 individuals for each of coronary heart disease, type 1 diabetes, type 2 diabetes, rheumatoid arthritis, Crohn's disease, bipolar disorder, and hypertension) and 3,000 shared controls.[15] This study was successful in uncovering many new disease genes underlying these diseases.[15][35]
Since these first landmark GWA studies, there have been two general trends.[36] One has been towards larger and larger sample sizes. In 2018, several genome-wide association studies are reaching a total sample size of over 1 million participants, including 1.1 million in a genome-wide study of educational attainment[37] follow by another in 2022 with 3 million individuals[38] and a study of insomnia containing 1.3 million individuals.[39] The reason is the drive towards reliably detecting risk-SNPs that have smaller effect sizes and lower allele frequency. Another trend has been towards the use of more narrowly defined phenotypes, such as blood lipids, proinsulin or similar biomarkers.[40][41] These are called intermediate phenotypes, and their analyses may be of value to functional research into biomarkers.[42]
A variation of GWAS uses participants that are first-degree relatives of people with a disease. This type of study has been named genome-wide association study by proxy (GWAX).[43]
A central point of debate on GWA studies has been that most of the SNP variations found by GWA studies are associated with only a small increased risk of the disease, and have only a small predictive value. The median odds ratio is 1.33 per risk-SNP, with only a few showing odds ratios above 3.0.[1][44] These magnitudes are considered small because they do not explain much of the heritable variation. This heritable variation is estimated from heritability studies based on monozygotic twins.[45] For example, it is known that 40% of variance in depression can be explained by hereditary differences, but GWA studies only account for a minority of this variance.[45]
Clinical applications and examples
A challenge for future successful GWA study is to apply the findings in a way that accelerates drug and diagnostics development, including better integration of genetic studies into the drug-development process and a focus on the role of genetic variation in maintaining health as a blueprint for designing new drugs and diagnostics.[46] Several studies have looked into the use of risk-SNP markers as a means of directly improving the accuracy of prognosis. Some have found that the accuracy of prognosis improves,[47] while others report only minor benefits from this use.[48] Generally, a problem with this direct approach is the small magnitudes of the effects observed. A small effect ultimately translates into a poor separation of cases and controls and thus only a small improvement of prognosis accuracy. An alternative application is therefore the potential for GWA studies to elucidate pathophysiology.[49]
Hepatitis C treatment
One such success is related to identifying the genetic variant associated with response to anti-hepatitis C virus treatment. For genotype 1 hepatitis C treated with Pegylated interferon-alpha-2a or Pegylated interferon-alpha-2b combined with ribavirin, a GWA study[50] has shown that SNPs near the human IL28B gene, encoding interferon lambda 3, are associated with significant differences in response to the treatment. A later report demonstrated that the same genetic variants are also associated with the natural clearance of the genotype 1 hepatitis C virus.[51] These major findings facilitated the development of personalized medicine and allowed physicians to customize medical decisions based on the patient's genotype.[52]
eQTL, LDL and cardiovascular disease
The goal of elucidating pathophysiology has also led to increased interest in the association between risk-SNPs and the gene expression of nearby genes, the so-called expression quantitative trait loci (eQTL) studies.[53] The reason is that GWAS studies identify risk-SNPs, but not risk-genes, and specification of genes is one step closer towards actionable drug targets. As a result, major GWA studies by 2011 typically included extensive eQTL analysis.[54][55][56] One of the strongest eQTL effects observed for a GWA-identified risk SNP is the SORT1 locus.[40] Functional follow up studies of this locus using small interfering RNA and gene knock-out mice have shed light on the metabolism of low-density lipoproteins, which have important clinical implications for cardiovascular disease.[40][57][58]
Atrial fibrillation
For example, a meta-analysis accomplished in 2018 revealed the discovery of 70 new loci associated with atrial fibrillation. It has been identified different variants associated with transcription factor coding-genes, such as TBX3 and TBX5, NKX2-5 o PITX2, which are involved in cardiac conduction regulation, in ionic channel modulation and cardiac development. It was also identified new genes involved in tachycardia (CASQ2) or associated with alteration of cardiac muscle cell communication (PKP2).[59]
Schizophrenia
Research using a High-Precision Protein Interaction Prediction (HiPPIP) computational model that discovered 504 new protein-protein interactions (PPIs) associated with genes linked to schizophrenia,.[60][61][62] While the evidence supporting the genetic basis of schizophrenia is not controversial, one study found that 25 candidate schizophrenia genes discovered from GWAS had little association with schizophrenia, demonstrating that GWAS alone may be insufficient to identify candidate genes.[63]
Conservation applications
Population level GWA studies may be used to identify adaptive genes to help evaluate ability of species to adapt to changing environmental conditions as the global climate becomes warmer.[64] This could help determine extirpation risk for species and could therefore be an important tool for conservation planning. Utilizing GWA studies to determine adaptive genes could help elucidate the relationship between neutral and adaptive genetic diversity.
Agricultural applications
Plant growth stages and yield components
GWA studies act as an important tool in plant breeding. With large genotyping and phenotyping data, GWAS are powerful in analyzing complex inheritance modes of traits that are important yield components such as number of grains per spike, weight of each grain and plant structure. In a study on GWAS in spring wheat, GWAS have revealed a strong correlation of grain production with booting data, biomass and number of grains per spike.[65] GWA study is also a success in study genetic architecture of complex traits in rice.[66]
Plant pathogens
The emergences of plant pathogens have posed serious threats to plant health and biodiversity. Under this consideration, identification of wild types that have the natural resistance to certain pathogens could be of vital importance. Furthermore, we need to predict which alleles are associated with the resistance. GWA studies is a powerful tool to detect the relationships of certain variants and the resistance to the plant pathogen, which is beneficial for developing new pathogen-resisted cultivars.[67]
Chicken
The first GWA study in chickens was done by Abasht and Lamont [68] in 2007. This GWA was used to study the fatness trait in F2 population found previously. Significantly related SNPs were found are on 10 chromosomes (1, 2, 3, 4, 7, 8, 10, 12, 15 and 27).
Limitations
GWA studies have several issues and limitations that can be taken care of through proper quality control and study setup. Lack of well defined case and control groups, insufficient sample size, control for population stratification are common problems.[2] On the statistical issue of multiple testing, it has been noted that "the GWA approach can be problematic because the massive number of statistical tests performed presents an unprecedented potential for false-positive results".[2] This is why all modern GWAS use a very low p-value threshold. In addition to easily correctible problems such as these, some more subtle but important issues have surfaced. A high-profile GWA study that investigated individuals with very long life spans to identify SNPs associated with longevity is an example of this.[69] The publication came under scrutiny because of a discrepancy between the type of genotyping array in the case and control group, which caused several SNPs to be falsely highlighted as associated with longevity.[70] The study was subsequently retracted,[71] but a modified manuscript was later published.[72] Now, many GWAS control for genotyping array. If there are substantial differences between groups on the type of genotyping array, as with any confounder, GWA studies could result in a false positive. Another consequence is that such studies are unable to detect the contribution of very rare mutations not included in the array or able to be imputed.[73]
Additionally, GWA studies identify candidate risk variants for the population from which their analysis is performed, and with most GWA studies historically stemming from European databases, there is a lack of translation of the identified risk variants to other non-European populations.[74] Alternative strategies suggested involve linkage analysis.[75][76] More recently, the rapidly decreasing price of complete genome sequencing have also provided a realistic alternative to genotyping array-based GWA studies. High-throughput sequencing does have potential to side-step some of the shortcomings of non-sequencing GWA.[77]
Fine-mapping
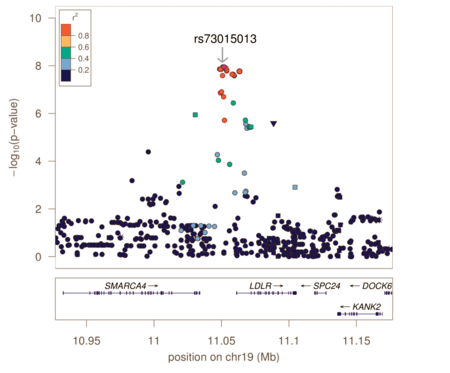
Genotyping arrays designed for GWAS rely on linkage disequilibrium to provide coverage of the entire genome by genotyping a subset of variants. Because of this, the reported associated variants are unlikely to be the actual causal variants. Associated regions can contain hundreds of variants spanning large regions and encompassing many different genes, making the biological interpretation of GWAS loci more difficult. Fine-mapping is a process to refine these lists of associated variants to a credible set most likely to include the causal variant.
Fine-mapping requires all variants in the associated region to have been genotyped or imputed (dense coverage), very stringent quality control resulting in high-quality genotypes, and large sample sizes sufficient in separating out highly correlated signals. There are several different methods to perform fine-mapping, and all methods produce a posterior probability that a variant in that locus is causal. Because the requirements are often difficult to satisfy, there are still limited examples of these methods being more generally applied.
See also
- Association mapping
- Transcriptome-wide association study
- Epidemiology
- Genetic diversity
- Gene–environment interaction
- Genomics
- Linkage disequilibrium
- Molecular epidemiology
- Polygenic score
- Population genetics
- Genetic epidemiology
- Common disease-common variant hypothesis
- Microbiome-wide association study
- Conservation biology
References
- ^ a b Manolio TA (July 2010). "Genomewide association studies and assessment of the risk of disease". The New England Journal of Medicine. 363 (2): 166–76. doi:10.1056/NEJMra0905980. PMID 20647212.
- ^ a b c Pearson TA, Manolio TA (March 2008). "How to interpret a genome-wide association study". JAMA. 299 (11): 1335–44. doi:10.1001/jama.299.11.1335. PMID 18349094.
- ^ "Genome-Wide Association Studies". National Human Genome Research Institute.
- ^ Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, et al. (December 2002). "Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction". Nature Genetics. 32 (4): 650–4. doi:10.1038/ng1047. PMID 12426569. S2CID 21414260.
- ^ Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. (April 2005). "Complement factor H polymorphism in age-related macular degeneration". Science. 308 (5720): 385–9. Bibcode:2005Sci...308..385K. doi:10.1126/science.1109557. PMC 1512523. PMID 15761122.
- ^ "GWAS Catalog: The NHGRI-EBI Catalog of published genome-wide association studies". European Molecular Biology Laboratory. Retrieved 18 April 2017.
- ^ a b c Bush WS, Moore JH (2012). Lewitter F, Kann M (eds.). "Chapter 11: Genome-wide association studies". PLOS Computational Biology. 8 (12): e1002822. Bibcode:2012PLSCB...8E2822B. doi:10.1371/journal.pcbi.1002822. PMC 3531285. PMID 23300413.
- ^ a b c Strachan T, Read A (2011). Human Molecular Genetics (4th ed.). Garland Science. pp. 467–495. ISBN 978-0-8153-4149-9.
- ^ "Online Mendelian Inheritance in Man". Archived from the original on 5 December 2011. Retrieved 6 December 2011.
- ^ a b Altmüller J, Palmer LJ, Fischer G, Scherb H, Wjst M (November 2001). "Genomewide scans of complex human diseases: true linkage is hard to find". American Journal of Human Genetics. 69 (5): 936–50. doi:10.1086/324069. PMC 1274370. PMID 11565063.
- ^ Risch N, Merikangas K (September 1996). "The future of genetic studies of complex human diseases". Science. 273 (5281): 1516–7. Bibcode:1996Sci...273.1516R. doi:10.1126/science.273.5281.1516. PMID 8801636. S2CID 5228523.
- ^ Greely HT (2007). "The uneasy ethical and legal underpinnings of large-scale genomic biobanks". Annual Review of Genomics and Human Genetics. 8: 343–64. doi:10.1146/annurev.genom.7.080505.115721. PMID 17550341.
- ^ The International HapMap Project, Gibbs RA, Belmont JW, Hardenbol P, Willis TD, Yu F, Yang H, Ch'Ang LY, Huang W (December 2003). "The International HapMap Project" (PDF). Nature. 426 (6968): 789–96. Bibcode:2003Natur.426..789G. doi:10.1038/nature02168. hdl:2027.42/62838. PMID 14685227. S2CID 4387110.
- ^ Schena M, Shalon D, Davis RW, Brown PO (October 1995). "Quantitative monitoring of gene expression patterns with a complementary DNA microarray". Science. 270 (5235): 467–70. Bibcode:1995Sci...270..467S. doi:10.1126/science.270.5235.467. PMID 7569999. S2CID 6720459.
- ^ a b c d Wellcome Trust Case Control Consortium, Burton PR (June 2007). "Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls". Nature. 447 (7145): 661–78. Bibcode:2007Natur.447..661B. doi:10.1038/nature05911. PMC 2719288. PMID 17554300.
- ^ a b c d Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT (February 2011). "Basic statistical analysis in genetic case-control studies". Nature Protocols. 6 (2): 121–33. doi:10.1038/nprot.2010.182. PMC 3154648. PMID 21293453.
- ^ Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. (September 2007). "PLINK: a tool set for whole-genome association and population-based linkage analyses". American Journal of Human Genetics. 81 (3): 559–75. doi:10.1086/519795. PMC 1950838. PMID 17701901.
- ^ Llinares-López F, Grimm DG, Bodenham DA, Gieraths U, Sugiyama M, Rowan B, Borgwardt K (June 2015). "Genome-wide detection of intervals of genetic heterogeneity associated with complex traits". Bioinformatics. 31 (12): i240-9. doi:10.1093/bioinformatics/btv263. PMC 4559912. PMID 26072488.
- ^ Ayati M, Erten S, Chance MR, Koyutürk M (December 2015). "MOBAS: identification of disease-associated protein subnetworks using modularity-based scoring". EURASIP Journal on Bioinformatics & Systems Biology. 2015 (1): 7. doi:10.1186/s13637-015-0025-6. PMC 5270451. PMID 28194175.
- ^ Ayati M, Koyutürk M (1 January 2015). "Assessing the Collective Disease Association of Multiple Genomic Loci". Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics. BCB '15. New York, NY, USA: ACM. pp. 376–385. doi:10.1145/2808719.2808758. ISBN 978-1-4503-3853-0. S2CID 5942777.
- ^ Marchini J, Howie B (July 2010). "Genotype imputation for genome-wide association studies". Nature Reviews Genetics. 11 (7): 499–511. doi:10.1038/nrg2796. PMID 20517342. S2CID 1465707.
- ^ Howie B, Marchini J, Stephens M (November 2011). "Genotype imputation with thousands of genomes". G3. 1 (6): 457–70. doi:10.1534/g3.111.001198. PMC 3276165. PMID 22384356.
- ^ Browning BL, Browning SR (February 2009). "A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals". American Journal of Human Genetics. 84 (2): 210–23. doi:10.1016/j.ajhg.2009.01.005. PMC 2668004. PMID 19200528.
- ^ Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (December 2010). "MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes". Genetic Epidemiology. 34 (8): 816–34. doi:10.1002/gepi.20533. PMC 3175618. PMID 21058334.
- ^ Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, Stephens M, Bustamante CD (November 2008). "Genes mirror geography within Europe". Nature. 456 (7218): 98–101. Bibcode:2008Natur.456...98N. doi:10.1038/nature07331. PMC 2735096. PMID 18758442.
- ^ Charney E (January 2017). "Genes, behavior, and behavior genetics". Wiley Interdisciplinary Reviews. Cognitive Science. 8 (1–2): e1405. doi:10.1002/wcs.1405. hdl:10161/13337. PMID 27906529.
- ^ Wittkowski KM, Sonakya V, Bigio B, Tonn MK, Shic F, Ascano M, Nasca C, Gold-Von Simson G (January 2014). "A novel computational biostatistics approach implies impaired dephosphorylation of growth factor receptors as associated with severity of autism". Translational Psychiatry. 4 (1): e354. doi:10.1038/tp.2013.124. PMC 3905234. PMID 24473445.
- ^ Barsh GS, Copenhaver GP, Gibson G, Williams SM (July 2012). "Guidelines for genome-wide association studies". PLOS Genetics. 8 (7): e1002812. doi:10.1371/journal.pgen.1002812. PMC 3390399. PMID 22792080.
- ^ Smith SM, Douaud G, Chen W, Hanayik T, Alfaro-Almagro F, Sharp K, Elliott LT (2021). "An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank". Nat Neurosci. 24 (5): 737–745. doi:10.1038/s41593-021-00826-4. PMC 7610742. PMID 33875891.
- ^ Sanna S, Li B, Mulas A, Sidore C, Kang HM, Jackson AU, et al. (July 2011). Gibson G (ed.). "Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability". PLOS Genetics. 7 (7): e1002198. doi:10.1371/journal.pgen.1002198. PMC 3145627. PMID 21829380.
- ^ Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA (June 2009). "Potential etiologic and functional implications of genome-wide association loci for human diseases and traits". Proceedings of the National Academy of Sciences of the United States of America. 106 (23): 9362–7. Bibcode:2009PNAS..106.9362H. doi:10.1073/pnas.0903103106. PMC 2687147. PMID 19474294.
- ^ Johnson AD, O'Donnell CJ (January 2009). "An open access database of genome-wide association results". BMC Medical Genetics. 10: 6. doi:10.1186/1471-2350-10-6. PMC 2639349. PMID 19161620.
- ^ Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA (April 2005). "Complement factor H variant increases the risk of age-related macular degeneration". Science. 308 (5720): 419–21. Bibcode:2005Sci...308..419H. doi:10.1126/science.1110359. PMID 15761120. S2CID 32716116.
- ^ Fridkis-Hareli M, Storek M, Mazsaroff I, Risitano AM, Lundberg AS, Horvath CJ, Holers VM (October 2011). "Design and development of TT30, a novel C3d-targeted C3/C5 convertase inhibitor for treatment of human complement alternative pathway-mediated diseases". Blood. 118 (17): 4705–13. doi:10.1182/blood-2011-06-359646. PMC 3208285. PMID 21860027.
- ^ "Largest ever study of genetics of common diseases published today" (Press release). Wellcome Trust Case Control Consortium. 6 June 2007. Retrieved 19 June 2008.
- ^ Ioannidis JP, Thomas G, Daly MJ (May 2009). "Validating, augmenting and refining genome-wide association signals". Nature Reviews Genetics. 10 (5): 318–29. doi:10.1038/nrg2544. PMC 7877552. PMID 19373277. S2CID 6463743.
- ^ Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linnér R, et al. (July 2018). "Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals". Nature Genetics. 50 (8): 1112–1121. doi:10.1038/s41588-018-0147-3. PMC 6393768. PMID 30038396.
- ^ Okbay A, Wu Y, Wang N, Jayashankar H, Bennett M, Nehzati SM, et al. (April 2022). "Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals". Nature Genetics. 54 (4): 437–449. doi:10.1038/s41588-022-01016-z. hdl:11368/3026010. PMC 9005349. PMID 35361970.
- ^ Jansen PR, Watanabe K, Stringer S, Skene N, Bryois J, Hammerschlag AR, et al. (March 2019). "Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways". Nature Genetics. 51 (3): 394–403. bioRxiv 10.1101/214973. doi:10.1038/s41588-018-0333-3. PMID 30804565.
- ^ a b c Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, et al. (January 2009). "Common variants at 30 loci contribute to polygenic dyslipidemia". Nature Genetics. 41 (1): 56–65. doi:10.1038/ng.291. PMC 2881676. PMID 19060906.
- ^ Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, et al. (October 2011). "Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes". Diabetes. 60 (10): 2624–34. doi:10.2337/db11-0415. PMC 3178302. PMID 21873549.
- ^ Danesh J, Pepys MB (November 2009). "C-reactive protein and coronary disease: is there a causal link?". Circulation. 120 (21): 2036–9. doi:10.1161/CIRCULATIONAHA.109.907212. PMID 19901186.
- ^ Liu JZ, Erlich Y, Pickrell JK (March 2017). "Case-control association mapping by proxy using family history of disease". Nature Genetics. 49 (3): 325–331. doi:10.1038/ng.3766. PMID 28092683. S2CID 5598845.
- ^ Ku CS, Loy EY, Pawitan Y, Chia KS (April 2010). "The pursuit of genome-wide association studies: where are we now?". Journal of Human Genetics. 55 (4): 195–206. doi:10.1038/jhg.2010.19. PMID 20300123.
- ^ a b Maher B (November 2008). "Personal genomes: The case of the missing heritability". Nature. 456 (7218): 18–21. doi:10.1038/456018a. PMID 18987709.
- ^ Iadonato SP, Katze MG (September 2009). "Genomics: Hepatitis C virus gets personal". Nature. 461 (7262): 357–8. Bibcode:2009Natur.461..357I. doi:10.1038/461357a. PMID 19759611. S2CID 7602652.

- ^ Muehlschlegel JD, Liu KY, Perry TE, Fox AA, Collard CD, Shernan SK, Body SC (September 2010). "Chromosome 9p21 variant predicts mortality after coronary artery bypass graft surgery". Circulation. 122 (11 Suppl): S60–5. doi:10.1161/CIRCULATIONAHA.109.924233. PMC 2943860. PMID 20837927.
- ^ Paynter NP, Chasman DI, Paré G, Buring JE, Cook NR, Miletich JP, Ridker PM (February 2010). "Association between a literature-based genetic risk score and cardiovascular events in women". JAMA. 303 (7): 631–7. doi:10.1001/jama.2010.119. PMC 2845522. PMID 20159871.
- ^ Couzin-Frankel J (June 2010). "Major heart disease genes prove elusive". Science. 328 (5983): 1220–1. Bibcode:2010Sci...328.1220C. doi:10.1126/science.328.5983.1220. PMID 20522751.

- ^ Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB (September 2009). "Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance". Nature. 461 (7262): 399–401. Bibcode:2009Natur.461..399G. doi:10.1038/nature08309. PMID 19684573. S2CID 1707096.
- ^ Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M (October 2009). "Genetic variation in IL28B and spontaneous clearance of hepatitis C virus". Nature. 461 (7265): 798–801. Bibcode:2009Natur.461..798T. doi:10.1038/nature08463. PMC 3172006. PMID 19759533.
- ^ Lu YF, Goldstein DB, Angrist M, Cavalleri G (July 2014). "Personalized medicine and human genetic diversity". Cold Spring Harbor Perspectives in Medicine. 4 (9): a008581. doi:10.1101/cshperspect.a008581. PMC 4143101. PMID 25059740.
- ^ Folkersen L, van't Hooft F, Chernogubova E, Agardh HE, Hansson GK, Hedin U, Liska J, Syvänen AC, Paulsson-Berne G, Paulssson-Berne G, Franco-Cereceda A, Hamsten A, Gabrielsen A, Eriksson P (August 2010). "Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease". Circulation: Cardiovascular Genetics. 3 (4): 365–73. doi:10.1161/CIRCGENETICS.110.948935. PMID 20562444.
- ^ Bown MJ, Jones GT, Harrison SC, Wright BJ, Bumpstead S, Baas AF, et al. (November 2011). "Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1". American Journal of Human Genetics. 89 (5): 619–27. doi:10.1016/j.ajhg.2011.10.002. PMC 3213391. PMID 22055160.
- ^ Coronary Artery Disease (C4D) Genetics Consortium (March 2011). "A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease". Nature Genetics. 43 (4): 339–44. doi:10.1038/ng.782. PMID 21378988. S2CID 39712343.
{{cite journal}}: CS1 maint: numeric names: authors list (link)
- ^ Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, et al. (December 2011). "Blood pressure loci identified with a gene-centric array". American Journal of Human Genetics. 89 (6): 688–700. doi:10.1016/j.ajhg.2011.10.013. PMC 3234370. PMID 22100073.
- ^ Dubé JB, Johansen CT, Hegele RA (June 2011). "Sortilin: an unusual suspect in cholesterol metabolism: from GWAS identification to in vivo biochemical analyses, sortilin has been identified as a novel mediator of human lipoprotein metabolism". BioEssays. 33 (6): 430–7. doi:10.1002/bies.201100003. PMID 21462369.

- ^ Bauer RC, Stylianou IM, Rader DJ (April 2011). "Functional validation of new pathways in lipoprotein metabolism identified by human genetics". Current Opinion in Lipidology. 22 (2): 123–8. doi:10.1097/MOL.0b013e32834469b3. PMID 21311327. S2CID 24020035.

- ^ Roselli C, Chafin M, Weng L (2018). "Multi-ethnic genome-wide association study for atrial fibrillation". Nature Genetics. 50 (9): 1225–1233. doi:10.1038/s41588-018-0133-9. PMC 6136836. PMID 29892015.
- ^ Ganapathiraju MK, Thahir M, Handen A, Sarkar SN, Sweet RA, Nimgaonkar VL, Loscher CE, Bauer EM, Chaparala S (27 April 2016). "Schizophrenia interactome with 504 novel protein-protein interactions". npj Schizophrenia. 2: 16012. doi:10.1038/npjschz.2016.12. PMC 4898894. PMID 27336055.
- ^ "New Schizophrenia Study Focuses on Protein-Protein Interactions". psychcentral.com. 3 May 2016. Archived from the original on 11 January 2020. Retrieved 22 April 2023.
- ^ Ganapathiraju M, Chaparala S, Lo C (April 2018). "F200. Elucidating The Role of Cilia in Neuropsychiatric Diseases Through Interactome Analysis". Schizophrenia Bulletin. 44 (suppl_1): S298-9. doi:10.1093/schbul/sby017.731. PMC 5887623.
- ^ Johnson EC, Border R, Melroy-Greif WE, de Leeuw CA, Ehringer MA, Keller MC (November 2017). "No Evidence That Schizophrenia Candidate Genes Are More Associated With Schizophrenia Than Noncandidate Genes". Biological Psychiatry. 82 (10): 702–708. doi:10.1016/j.biopsych.2017.06.033. PMC 5643230. PMID 28823710.
- ^ Willi Y, Kristensen TN, Sgrò CM, Weeks AR, Ørsted M, Hoffmann AA (January 2022). "Conservation genetics as a management tool: The five best-supported paradigms to assist the management of threatened species". Proceedings of the National Academy of Sciences of the United States of America. 119 (1): e2105076119. doi:10.1073/pnas.2105076119. PMC 8740573. PMID 34930821.
- ^ Turuspekov Y, Baibulatova A, Yermekbayev K, Tokhetova L, Chudinov V, Sereda G, et al. (November 2017). "GWAS for plant growth stages and yield components in spring wheat (Triticum aestivum L.) harvested in three regions of Kazakhstan". BMC Plant Biology. 17 (Suppl 1): 190. doi:10.1186/s12870-017-1131-2. PMC 5688510. PMID 29143598.
- ^ Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, et al. (September 2011). "Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa". Nature Communications. 2 (1): 467. Bibcode:2011NatCo...2..467Z. doi:10.1038/ncomms1467. PMC 3195253. PMID 21915109.
- ^ Bartoli C, Roux F (2017). "Genome-Wide Association Studies In Plant Pathosystems: Toward an Ecological Genomics Approach". Frontiers in Plant Science. 8: 763. doi:10.3389/fpls.2017.00763. PMC 5441063. PMID 28588588.
- ^ Abasht B, Lamont SJ (October 2007). "Genome-wide association analysis reveals cryptic alleles as an important factor in heterosis for fatness in chicken F2 population". Animal Genetics. 38 (5): 491–498. doi:10.1111/j.1365-2052.2007.01642.x. PMID 17894563.
- ^ Sebastiani P, Solovieff N, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Perls TT (July 2010). "Genetic signatures of exceptional longevity in humans". Science. 2010 (5987). doi:10.1126/science.1190532. PMID 20595579. (Retracted)

- ^ MacArthur D (8 July 2010). "Serious flaws revealed in "longevity genes" study". Wired. Retrieved 7 December 2011.
- ^ Sebastiani P, Solovieff N, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Perls TT (July 2011). "Retraction". Science. 333 (6041): 404. doi:10.1126/science.333.6041.404-a. PMID 21778381.

- ^ Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Hoh J, Perls TT (18 January 2012). "Genetic signatures of exceptional longevity in humans". PLOS ONE. 7 (1): e29848. Bibcode:2012PLoSO...729848S. doi:10.1371/journal.pone.0029848. PMC 3261167. PMID 22279548.
- ^ Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D (May 2019). "Benefits and limitations of genome-wide association studies". Nature Reviews Genetics. 20 (8): 467–484. doi:10.1038/s41576-019-0127-1. ISSN 1471-0056. PMID 31068683. S2CID 148570302.
- ^ Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M (May 2010). "Genome-wide association studies in diverse populations". Nature Reviews Genetics. 11 (5): 356–66. doi:10.1038/nrg2760. PMC 3079573. PMID 20395969.
- ^ Sham PC, Cherny SS, Purcell S, Hewitt JK (May 2000). "Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data". American Journal of Human Genetics. 66 (5): 1616–30. doi:10.1086/302891. PMC 1378020. PMID 10762547.
- ^ Borecki IB (2006). "Linkage and Association Studies". Encyclopedia of Life Sciences. John Wiley & Sons, Ltd. doi:10.1038/npg.els.0005483. ISBN 9780470015902.
{{cite book}}:|work=ignored (help) - ^ Visscher PM, Goddard ME, Derks EM, Wray NR (May 2012). "Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses". Molecular Psychiatry. 17 (5): 474–85. doi:10.1038/mp.2011.65. PMID 21670730.

External links
- Genotype-phenotype interaction software tools and databases on omicX[permanent dead link]
- Statistical Methods for the Analysis of Genome-Wide Association Studies [video lecture series]
- Whole genome association studies — by the National Human Genome Research Institute
- GWAS Central — a central database of summary-level genetic association findings
- Barrett J (18 July 2010). "How to read a genome-wide association study". Genomes Unzipped.
- Consortia of genome-wide association studies (GWAS) Archived 26 February 2018 at the Wayback Machine — by Bennett SN, Caporaso, NE, et al.
- PLINK — whole genome association analysis toolset
- ENCODE threads explorer Impact of functional information on understanding variation. Nature (journal)
