Monocercomonoides
This article may be too technical for most readers to understand. (May 2019) |
| Monocercomonoides | |
|---|---|
| |
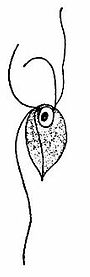
| Monocercomonoides melolanthae | |
| Scientific classification | |
| Domain: | |
| (unranked): | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | Monocercomonoides Travis, 1932
|
| Species | |
| |
Monocercomonoides is a genus of flagellate Excavata belonging to the order Oxymonadida. It was established by Bernard V. Travis and was first described as those with "polymastiginid flagellates having three anterior flagella and a trailing one originating at a single basal granule located in front of the anteriorly positioned nucleus, and a more or less well-defined axostyle".[14] It is the first eukaryotic genus to be found to completely lack mitochondria, and all hallmark proteins responsible for mitochondrial function. The genus also lacks any other mitochondria related organelles (MROs) such as hydrogenosomes or mitosomes.[15] Data suggests that the absence of mitochondria is not an ancestral feature, but rather due to secondary loss. Monocercomonoides sp. was found to obtain energy through an enzymatic action of nutrients absorbed from the environment.[15] The genus has replaced the iron-sulfur cluster assembly pathway with a cytosolic sulfur mobilization system, likely acquired by horizontal gene transfer from a eubacterium of a common ancestor of oxymonads.[16] These organisms are significant because they undermine assumptions that eukaryotes must have mitochondria to function properly. The genome of Monocercomonoides exilis has approximately 82 million base pairs (82 Mbp), with 18 152 predicted protein-coding genes.[17]
Habitat and ecology
[edit]Most Monocercomonoides species are obligate animal symbionts that live in the digestive tracts of insects, amphibians, reptiles, and mammals.[18] Monocercomonoides are common in insect orders Orthoptera and Coleoptera. The species Monocercomonoides qadrii are found in the rectum of the larva of the dung-beetle (Oryctes rhinoceros).[19] M. caviae, M. wenrichi, M. quadrifunilis, and M. exilis are found in the caecum of guinea pigs, and M. caprae has been found in the rumen of goats.[20] Interestingly, some Monocercomonoides species were isolated from cesspits,[21] suggesting that they might be able to survive outside of the host in certain environmental conditions. The organism uses enzymes found in its cytoplasm to break down food and furnish energy since there is no mitochondria or oxygen presence.[22] It has been noted that Monocercomonoides ingest bacteria or wood and feed by pinocytosis, however, limited studies have been done on feeding style.
Morphology
[edit]Monocercomonoides are small free-swimming, single-cell organisms ranging from 5-12μm in length, and 4.5-14.5μm in width.[19] The body may be ovoidal, pyriform, spherical or subspherical; however, they lack holdfasts and have small axostyles.[23] The axostyle is a single, contractible appendage made of microtubules that originates from the posterior end of the preaxostyle, and is situated near the posterior pair of the basal bodies (known as blepharoplast in older cytological literature).[23] The cytoskeleton is based around four basal bodies, an anterior pair and a posterior pair.[24] The preaxostyle runs between the two pairs of basal bodies and is composed of a broad, curved sheet of microtubules.[24] The inner face of the microtubule sheet adheres to a paracrystalline fibre (about 50 nm thick) which is a common characteristic of all oxymonads.[24] Monocercomonoides sp. has four flagella that originate in two pairs and arise from each basal body found in the anterior end.[23] Three of the four flagella are roughly equal in length (9.5-18μm) and the fourth trailing flagellum is slightly longer, measuring between 9.0 and 24.5μm.[19] The flagella have a beating action and are used for rapid movement. The proximal part of the long flagellum may adhere to the pellicle, which causes it to trail posteriorly.[23] The trailing flagellum is always directed backwards and is attached to the body for a considerable distance (6-9μm) by an accessory filament called a funis.[19] There are one to four filaments (rib-like strictures) extending backwards beneath the body surface.[19] In some parasites, the flagella end in acronemes. The nucleus is generally situated near the anterior end of the body and contains a central endosome surrounded by chromatin granules, some species have pelta-like structures below the nucleus.[23] The cytoplasm is granular with or without vacuoles.[23] Electron microscopic imaging of Monocercomonoides has found that the intracellular morphology lacks any Golgi apparatus, mitochondria, or potential homologs of the two; Golgi-associated proteins have been detected, but mitochondrial ones have not.[15]
Metabolic processes
[edit]Monocercomonoides sp. strain PA203 (later described as M. exilis[21]) is the first eukaryote discovered to lack any trace of mitochondria. In all other eukaryotes that seemingly lack mitochondria, there is nuclear DNA that contains some of the genes required to assemble mitochondria, but no such genes are present in Monocercomonoides.[15] It also lacks any genes ordinarily found in mitochondrial DNA, and genes used to make the energy-extracting enzymes present in mitochondria. Monocercomonoides are able to get some energy from glucose using anaerobic metabolic pathways that operate in the cytoplasm, however, most of its energy is obtained using enzymes that break down the amino acid arginine.[24]
Glycolytic pathway
[edit]Each molecule of glucose catabolized in Monocercomonoides yields less ATP compared to mitochondrial eukaryotes that use the tricarboxylic acid cycle and electron transport chain.[25] To aid in energy conservation, Monocercomonoides has adapted alternative glycolytic enzymes. Four alternative glycolytic enzymes include pyrophosphate-fructose-6-phosphate phosphotransferase (PFP), fructose-bisphosphate aldolase class II (FBA class II), 2,3-bisphosphoglycerate independent phosphoglycerate mutase (iPGM), and pyruvate phosphate dikinase (PPDK).[25] Glucose-6-phosphate isomerase (GPI) is predicted to be in Monocercomonoides since it is universally distributed among Eukaryotes, Bacteria, and some Archaea and essential in catabolic glycolysis, but has not yet been found.[25] Most of the glycolytic enzymes are the standard eukaryotic versions, making Monocercomonoides' metabolic pathway a mosaic similar to that of other anaerobes.[25]
The addition of PPDK to the conversion of phosphoenolpyruvate to pyruvate (typically catalyzed solely by pyruvate kinase) has a strong effect on ATP conservation.[25] Both PFP and PPDK rely on inorganic phosphate (PPi) as the phosphate donor; therefore rather than hydrolyzing ATP, the ATP yield is increased by using a by-product of the cell's anabolic processes as an energy source.[25] These reactions are able to allow for greater ATP conservation and regulation of glycolysis due to the PPDK's reversible nature and use of inorganic phosphate where pyruvate kinase only catalyzes the forward reaction.[25]
Arginine deiminase pathway
[edit]In addition to the adjusted glycolysis, Monocercomonoides contain enzymes needed in the arginine deiminase (degradation) pathway.[15] The arginine deiminase pathway may be used for ATP production, as in Giardia intestinalis and Trichomonas vaginalis.[15] In G. intestinalis (an anaerobic unicellular eukaryote) this pathway produces eight times more ATP than sugar metabolism, and a similar output is expected in Monocercomonoides, but has yet to be confirmed.[15] All 3 enzymes of the arginine deiminase pathway are localized in the cytosol of Monocercomonoides exilis which may reflect an ancestral state in Metamonada.[26]
Iron-sulfur cluster
[edit]Iron-sulfur clusters are important protein components that are synthesized by mitochondria.[16] The main function of these small inorganic prosthetic groups is mediating electron transport, which makes them a key part of photosynthesis, respiration, DNA replication/repair, and regulation of gene expression.[16] In eukaryotic cells, the common pathway for Fe-S cluster synthesis is ISC (iron-sulfur cluster). In the cytosol, a cytosolic iron-sulfur cluster assembly (CIA) forms Fe-S cluster-containing proteins that are responsible for the maturation of nuclear Fe-S proteins. CIA is unique to eukaryotes and does not have prokaryotic homologs.[16] The mitochondrial ISC pathway is believed to be necessary for the function of CIA since it synthesizes and transports uncharacterized sulfur-containing precursor to the cytosol, and is a major reason for retention of mitochondrial-related organelles in anaerobic eukaryotes.[16] The genus Monocercomonoides contains the CIA pathways but completely lacks the ISC pathway, along with any mitochondrial proteins.[16] The genus contains a reduced version of the SUF (sulfur utilization factor) pathway, having only three proteins - SufB, SufC, and SufU.[16] The SUF pathway is a known pathway of prokaryotes, and it is believed that the genes used to build Monocercomonoides' SUF system had to have come from prokaryotes.[16] However, Monocercomonoides' SUF proteins were found to not be related to plastid homologues, or any other microbial eukaryotes.[16] It was proposed that the pathway was acquired from a eubacterium by horizontal gene transfer (HGT) in the common ancestor of Monocercomonoides and Paratrismastrix (a sister taxon of oxymonads).[16] The genetic acquisition has not been demonstrated despite the assumption that it must have occurred.
Mitochondrial acquisition and loss
[edit]Monocercomonoides contain no detectable sign that mitochondria were ever part of the organism.[15] However, since it is widely accepted that all eukaryotes have a common ancestor that evolved mitochondria, it is believed that mitochondria must have once been present in the ancestors to oxymonads and then secondarily lost. The amitchondrial genus demonstrates that mitochondria are not absolutely essential for life of a eukaryotic cell.
Genomic structure
[edit]The lack of mitochondria or any mitochondria-related organelles in Monocercomonoides exilis is confirmed by its genome sequence. A complete genome sequence analysis of Monocercomonoides exilis strain PA203 from Chinchilla lanigera was conducted.[15] The estimated size of the genome is ~75Mb and the number of predicted protein-coding genes is 16,629.[15] A more recent re-sequencing of the genome using Oxford nanopore showed that the genome is ~82 Mbp in size.[17] Homology searches reveal a lack of genes that encode mitochondrial import machinery, metabolite transport proteins, and iron-sulfur clusters.[15][17] Additionally, an absence of targeted important genes and genes coding for mitochondrial membrane proteins were revealed when a search for specific N-terminal and C-terminal sequences was conducted.[15][17] Genes that are typically encoded on mitochondrial genomes (mtDNA) were not found among the assembled scaffold, suggesting Monocercomonoides lacks mtDNA.[15] 18S RNA genes were sequenced and found to be 2,927 nt long, and is among the longest known.[15][21] Some expansions were specific to Monocercomonoides, but many were similar to those in other oxymonad genera but substantially longer.[15] Comparisons of genes coding for 𝛼-tubulin, 𝛽-tubulin, 𝛾-tubulin, EF-1𝛼, EF-2, cytHSP70, ubiquitin, 18S rRNA, and HSP90 allow the placement of oxymonads near diplomonads and trichomonads, with Monocercomonoides and Streblomastix making up the oxymonad branch.[15]
References
[edit]- ^ a b Mali, M.; Kulkarni, S.; Mali, S. (2001). "Two species of flagellates of the genus Monocercomonoides Travis, 1932 from the gut of dung beetle larva (Oryctes rhinoceros) in India". Geobios (Jodhpur). 28 (4): 201–204.
- ^ Mali, M.; Patil, D. (2003). "The morphology of Monocercomonoides aurangabadae n. sp. a flagellata from the gut of Blatta germanica". Uttar Pradesh Journal of Zoology. 23 (2): 117–119.
- ^ Jensen, E.A.; Hammond, D.M. (1964). "A morphological study of trichomonads and related flagellates from the bovine digestive tract". Journal of Protozoology. 11 (3): 386–394. doi:10.1111/j.1550-7408.1964.tb01768.x. PMID 14207121.
- ^ Krishnamurthy, R.; Madre, V.E. (1979). "Studies on two flagellates of the genus Monocercomonoides Travis, 1932 (Mastigophora: Polymastigina) from amphibians and reptiles in India". Acta Protozoologica. 18 (2): 251–257.
- ^ a b c Rao, T.B. (1969). "The morphology and incidence of the genus Monocercomonoides (Grassi, 1879) Travis, 1932, of insects found in Andhra Pradesh, India". Proceedings of the Indian Academy of Sciences, Section B. 70 (5): 208–214. doi:10.1007/BF03052226. S2CID 127320109. Retrieved 11 February 2018.
- ^ Sultana, T.; Krishnamurthy, R. (1978). "Monocercomonoides gryllusae n. sp. (Mastigophora: Oxymonadida) from Gryllus bimaculatus". Geobios (Jodhpur). 5 (3): 114–115.
- ^ Radek, R. (1994). "Monocercomonoides termitis n. sp., an oxymonad from the lower termite Kalotermes sinaicus". Archiv für Protistenkunde. 144 (4): 373–382. doi:10.1016/S0003-9365(11)80240-X.
- ^ Radek, R. (1997). "Monocercomonoides hausmanni nom. nov, a New Species Name for M. termitis Radek, 1994". Archiv für Protistenkunde. 147 (3–4): 411. doi:10.1016/S0003-9365(97)80068-1.
- ^ Mali, M.; Mali, S. (2004). "The Monocercomonoides khultabadae n.sp., a new flagellate from the gut of Pycnoscelus surinamensis". Uttar Pradesh Journal of Zoology. 24 (1): 55–58.
- ^ a b Krishnamurthy, R.; Sultana, T. (1976). "The morphology of two new flagellates of the genus Monocercomonoides Travis, 1932 from insects in India". Proceedings of the Indian Academy of Sciences, Section B. 84 (3): 109–115. doi:10.1007/BF03045588. S2CID 104326570. Retrieved 11 February 2018.
- ^ a b Krishnamurthy, R. (1967). "Two new species of the genus Monocercomonoides Travis, 1932 (protozoa: Mastigophora) from reptiles". Proceedings of the Indian Academy of Sciences, Section B. 66 (5): 184–191. doi:10.1007/BF03052183. S2CID 83345325. Retrieved 11 February 2018.
- ^ Abraham, R. (961). "A description of Monocercomonoides sayeedi n. sp., from the rumen of an Indian goat". Zeitschrift für Parasitenkunde. 20 (6): 558–562. doi:10.1007/BF00333238. S2CID 26344904.
- ^ Krishnamurthy, R.; Sultana, T. (1979). "A new flagellate of the genus Monocercomonoides Travis, 1932 from a termite". Proceedings of the National Academy of Sciences, India Section B. 49 (2): 85–87.
- ^ Travis, B. V. 1932. A Discussion of Synonymy in the Nomenclature of Certain Insect Flagellates, with the Description of a New Flagellate from the Larvae of Ligyrodes relictus Say (Coleoptera-Scarabaeidae). Iowa State Coll J. Sci., 6, 317–323.
- ^ a b c d e f g h i j k l m n o p Karnkowska, Anna; Vacek, Vojtěch; Zubáčová, Zuzana; Treitli, Sebastian C.; Petrželková, Romana; Eme, Laura; Novák, Lukáš; Žárský, Vojtěch; Barlow, Lael D.; Herman, Emily K.; Soukal, Petr (2016). "A Eukaryote without a Mitochondrial Organelle". Current Biology. 26 (10): 1274–1284. Bibcode:2016CBio...26.1274K. doi:10.1016/j.cub.2016.03.053. PMID 27185558. S2CID 3933236.
- ^ a b c d e f g h i j Vacek, V., Novak, L. V. F., Treitli, S. C., et al. 2018. Fe–S Cluster Assembly in Oxymonads and Related Protists. Molecular Biology and Evolution. 35(11): 2712-2718.
- ^ a b c d Treitli, Sebastian Cristian; Peña-Diaz, Priscila; Hałakuc, Paweł; Karnkowska, Anna; Hampl, VladimírYR 2021 (2021). "High quality genome assembly of the amitochondriate eukaryote Monocercomonoides exilis". Microbial Genomics. 7 (12): 000745. doi:10.1099/mgen.0.000745. ISSN 2057-5858. PMC 8767320. PMID 34951395.
{{cite journal}}: CS1 maint: numeric names: authors list (link) - ^ Hampl, Vladimir (2017), Archibald, John M.; Simpson, Alastair G.B.; Slamovits, Claudio H. (eds.), "Preaxostyla", Handbook of the Protists, Cham: Springer International Publishing, pp. 1139–1174, doi:10.1007/978-3-319-28149-0_8, ISBN 978-3-319-28147-6, retrieved 2022-08-01
- ^ a b c d e Bhaskar Roa, T. 1969. The morphology and Incidence of the genus Monocercomonoides of insects found in Andhra Pradesh, India. Proceedings of the Indian Academy of Sciences. 70(5): 208-214.
- ^ Flynn, F. J. 1923. Parasites of Laboratory Animals. Iowa: Blackwell Publishing.
- ^ a b c Treitli, Sebastian C.; Kotyk, Michael; Yubuki, Naoji; Jirounková, Eliška; Vlasáková, Jitka; Smejkalová, Pavla; Šípek, Petr; Čepička, Ivan; Hampl, Vladimír (2018-11-01). "Molecular and Morphological Diversity of the Oxymonad Genera Monocercomonoides and Blattamonas gen. nov". Protist. 169 (5): 744–783. doi:10.1016/j.protis.2018.06.005. ISSN 1434-4610. PMID 30138782. S2CID 52077969.
- ^ Leslie, M. 2016. First eukaryotes found without a normal cellular power supply. Science Mag.
- ^ a b c d e f Laird, M. 1955. Intestinal Flagellates from Some New Zealand Insects. Transactions and Proceedings of the Royal Society of New Zealand. 84: 297-307.
- ^ a b c d Simpson, A. G. B., Radek, R., Dacks, J. B., and O'Kelly, C. J. 2002. How oxymonads lost their groove: An ultrastructural comparison of Monocercomonoides and excavate taxa. Journal of Eukaryotic Microbiology 49: 239-248.
- ^ a b c d e f g Liapounova, N. A., Hampl, V., Gordon, P. M., Sensen, C.W., Gedamu, L., and Dacks, J.B. 2006. Reconstructing the mosaic glycolytic pathway of the anaerobic eukaryote Monocercomonoides. Eukaryotic Cell 5(12): 2138-2146.
- ^ Novák, Lukáš; Zubáčová, Zuzana; Karnkowska, Anna; Kolisko, Martin; Hroudová, Miluše; Stairs, Courtney W.; Simpson, Alastair G. B.; Keeling, Patrick J.; Roger, Andrew J.; Čepička, Ivan; Hampl, Vladimír (2016-10-06). "Arginine deiminase pathway enzymes: evolutionary history in metamonads and other eukaryotes". BMC Evolutionary Biology. 16 (1): 197. Bibcode:2016BMCEE..16..197N. doi:10.1186/s12862-016-0771-4. ISSN 1471-2148. PMC 5052871. PMID 27716026.
External links
[edit] Data related to Monocercomonoides at Wikispecies
Data related to Monocercomonoides at Wikispecies
