RBM10
RNA-binding motif 10 is a protein that is encoded by the RBM10 gene.[5][6][7][8] This gene maps on the X chromosome at Xp11.23 in humans. RBM10 is a regulator of alternative splicing.[9][10][11] Alternative splicing is a process associated with gene expression to produce multiple protein isoforms from a single gene, thereby creating functional diversity and cellular complexity.[12] RBM10 influences the expression of many genes,[9][10][13][14][15] participating in various cellular processes and pathways such as cell proliferation and apoptosis.[10][16] Its mutations are associated with various human diseases[17][18][19][20][21][22] such as TARP syndrome,[22][17] an X-linked congenital disorder in males resulting in pre‐ or postnatal lethality, and various cancers in adults.[18][19]
Gene and protein[edit]
The RBM10 gene spans ~41.6 kb and contains 24 exons. This gene is subjected to X-inactivation,[6][7] in which one of the two RBM10 genes in female cells is transcriptionally silenced by heterochromatin formation.
RBM proteins constitute a large family of RNA-binding proteins (RBPs). There are 52 RBM proteins (HGNC: HUGO Gene Nomenclature Committee), each containing one to several RNA-binding domains called RNA recognition motifs (RRMs). RBM10 contains two RRMs (RRM1 and RRM2) and other domains such as two zinc fingers (ZnFs), an octamer repeat (OCRE), three nuclear localization signals (NLSs), and a glycine-rich domain (G-patch). The amino acid (aa) sequence of RBM10 is conserved among mammals. Human RBM10 isoform 1 shares 96% and 97% sequence homology with those of mice and rats, respectively, indicating that the molecular functions of RBM10 are essentially the same in humans and rodents.
RBM10 has multiple isoforms, generated via alternative splicing events of the RBM10 primary transcript. The main isoforms, 1–4, may contain an exon 4 sequence (77 residues) and/or a Val residue corresponding to the last codon of exon 10. Isoform 1 (930 residues) contains both the exon 4 sequence and V354, whereas isoform 4 (929 residues) does not contain this valine residue. Similarly, the exon 4–minus isoform 3 (853 residues) contains V277, whereas isoform 2 (852 residues) does not. Isoform 5 (995 residues) has a longer 65-aa N-terminus, compared with that of isoform 1. In addition, automated computational analysis using the Gnomon gene prediction tool (NCBI gene) has shown that there may be more than 10 different RBM isoforms.
Function[edit]
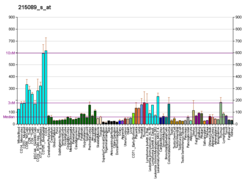
RBM10 is ubiquitously expressed in almost every type of cell, both growing as well as quiescent (UniProtKB-P98175 [human] and Q99KG3 [mouse]; The Human Protein Atlas). In general, it is more strongly expressed in actively transcribing cells.[23]
In the alternative splicing regulation, RBM10 promotes the exclusion of an exon, called a cassette or alternative exon, from target pre-mRNAs, and less frequently, other alternative splicing events such as alternative 5ʹ-splice site selection.[9][10][11][24] In the exon skipping process, RBM10 binds close to the 3ʹ- and 5ʹ-splice sites of cassette exons and interferes with the recognition and/or the pairing of the splice sites, thereby enhancing the pairing of the splice sites distal to the cassette exons, which ultimately leads to the exclusion of the exons together with the flanking upstream and downstream introns.[9][10][24]
The diversity of target RNAs bound by RBM10 in cells suggests that it is involved in various metabolic processes such as oxidative phosphorylation; pathways linked to cell proliferation, apoptosis, cell adhesion, and actin/cytoskeleton reorganization; and various diseases such as cancers and neurodegenerative diseases.[10][16][25] These data, together with the ubiquitous expression of RBM10, indicate that it is a fundamental cellular component participating in various cellular processes. In addition to alternative splicing regulation, RBM10 participates in other reactions. Some examples are polyadenylation of cardiac pre-mRNAs of anti-hypertrophy regulators, wherein it acts as a co-regulator of STAR-poly(A) polymerase,[26] stabilization of angiotensin II receptor mRNA by binding to its 3ʹ-UTR,[27] let-7g miRNA biogenesis through interaction with its precursor,[28] p53 stabilization by binding to its negative regulator, MDM2,[29] cell cycle arrest,[30][31] and anti-viral reactions.[32]
RBM10 localizes to the nucleoplasm, where transcription and splicing occur, as well as in membrane-less nuclear compartments called S1-1 nuclear bodies (S1-1 NBs).[23] The numbers (ca. 10–40 per nucleus) and sizes (ca. 0.5 µm) of S1-1 NBs vary with the cell type and cellular conditions. When RNA polymerase II transcription decreases, RBM10 in the nucleoplasm is sequestered in S1-1 NBs, which become larger and spherical; when transcription is restored, RBM10 and the S1-1 NBs return to their initial states.[23] S1-1 NBs often overlap with nuclear speckles (also known as splicing speckles or interchromatin granule clusters),[23][33] seemingly indicating a close functional relationship between these nuclear domains, i.e., alternative splicing regulation and splicing reaction.
Regulation[edit]
In females, most genes on one of the two X chromosomes are transcriptionally silenced by heterochromatin formation, and RBM10 is subjected to this X-inactivation.[6][7][34] In addition, there are mechanisms to control elevated cellular levels of RBM10. RBM10 auto-regulates its overexpressed pre-mRNA by alternative splicing to exclude exon 6 or 12, which generates a premature stop codon in the transcripts, leading to their degradation through nonsense-mediated mRNA decay (NMD).[14] When RNA polymerase II transcription decreases, RBM10 is sequestered in S1-1 NBs until transcription is restored.[23] In addition, RBM10 undergoes post-translational modifications: phosphorylation at many sites in response to various stimuli and changes in cellular conditions (UniProtKB-P98175; PhosphoSitePlus RBM10), as well as ubiquitylation,[35][36] acetylation,[37] and methylation.[38] However, the molecular and biological significance of these various post-translational modifications of RBM10 is not well understood.
Clinical significance[edit]
Mutations in RBM10 are associated with various human diseases. The phenotypes caused by RBM10 mutations differ by the stages of development and affected tissues. Typical examples are TARP syndrome, an X-linked pleiotropic developmental malformation in neonates,[17][22] and various cancers such as lung adenocarcinoma (LUAD)[18] and bladder carcinoma (BLCA) in adults.[19] These diseases are more common in males than in females.[39][40][41] One reason for this is the difference in the copy number of the RBM10 gene in a cell (one in male cells and two in female cells). Mutations in RBM10 occur throughout the molecule, and many of them are null mutations. TARP syndrome is generally pre- or postnatally lethal.[17][42][43] However, patients aged 11, 14, and 28 years have been reported to escape these null mutations.[44][9][45] RBM10 mutations have also been identified in other cancers[46] such as renal carcinomas,[47][48][49] pancreatic cancers,[50][51] colorectal cancers,[52][53] thyroid cancers,[54][55][56] breast cancers,[57] bile duct cancers,[58][59] prostate cancer,[57] and brain tumor meningiomas and astroblastomas.[60][61]
NUMB is the most studied downstream effector of RBM10. RBM10 promotes the skipping of exon 9 of the NUMB transcript, producing a NUMB isoform that causes ubiquitination followed by proteasomal degradation of the Notch receptor, and thereby inhibits the Notch signaling cell-proliferation pathway.[10][62][20] In various cancers, RBM10 mutations that inactivate or reduce its alternative splicing regulatory activity enhance the production of the exon 9–including NUMB isoform, which promotes cancer cell proliferation through the Notch pathway.[10][63][64]
RBM10 suppresses cell proliferation[10][27][63][64][65][66][29] and promotes apoptosis.[27][64][65][29][67][68] Hence, it is generally regarded as a tumor suppressor. However, in certain cases, it may exert an opposite oncogenic function by acting as a tumor promoter or growth enhancer,[16][69][70] presumably due to the cellular contexts composed of different constituents and active pathways. A typical example of this is patients with pancreatic ductal adenocarcinoma (PDAC) having RBM10 mutations, who exhibit a survival rate remarkably higher than the general 5-year PDAC survival rate of less than 7–8%.[50][71][72]
Paralogs and splicing network[edit]
RBM5 and RBM6 are paralogs of RBM10. They were generated by gene duplications during genome evolution. They generally function as tumor suppressors,[10][73][74][75][76][77][78][79] and their mutations are often identified in lung cancers.[21] RBM5, RBM6, and RBM10 regulate alternative splicing[10][80][81] and generally act on different RNAs; however, in certain cases, they act on the same subset of RNAs, likely producing synergistic or antagonistic effects.[10] There is a cross-regulation between RBM5 and RBM10; RBM10 lowers RBM5 transcript levels by alternative splicing–coupled NMD.[14] Furthermore, RBM10 perturbation (knockdown or overexpression) brings about splicing alterations in multiple splicing regulators, including RBM5, and also significantly influences the expression of other splicing regulators, including RBM10 itself.[9][14] In addition, RBM10 primary transcripts are subjected to alternative splicing at several exons by unidentified splicing regulators, leading to the generation of multiple RBM10 isoforms. These data suggest the existence of an alternative splicing network formed by RBM5, RBM6, and RBM10, as well as other splicing regulators.[82] Studies on such networks are expected to promote our understanding of transcriptomic homeostasis regulated by splicing and the molecular and biological significance of RBM10 in cells.
RBM10 regulates hundreds of genes.[9][10][13][14][15] Further studies on the various RBM10-mediated processes and pathways may help elucidate the pathogenesis and progression of diseases caused by RBM10 mutations and the mechanisms of the antithetical actions of RBM10 as a tumor suppressor, and in certain cases, a tumor promoter, and provide clues for better treatment of the diseases.
Notes[edit]
The 2021 version of this article was updated by an external expert under a dual publication model. The corresponding academic peer reviewed article was published in Gene and can be cited as: Akira Inoue, Akira Inoue (30 May 2021). "RBM10: Structure, functions, and associated diseases". Gene. Gene Wiki Review Series. doi:10.1016/J.GENE.2021.145463. ISSN 0378-1119. PMID 33515724. Wikidata Q108806632. |
References[edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000182872 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000031060 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N (August 1995). "Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1". DNA Research. 2 (4): 167–74, 199–210. doi:10.1093/dnares/2.4.167. PMID 8590280.
- ^ a b c Coleman MP, Ambrose HJ, Carrel L, Németh AH, Willard HF, Davies KE (January 1996). "A novel gene, DXS8237E, lies within 20 kb upstream of UBE1 in Xp11.23 and has a different X inactivation status". Genomics. 31 (1): 135–8. doi:10.1006/geno.1996.0022. PMID 8808293.
- ^ a b c Thiselton DL, McDowall J, Brandau O, Ramser J, d'Esposito F, Bhattacharya SS, et al. (April 2002). "An integrated, functionally annotated gene map of the DXS8026-ELK1 interval on human Xp11.3-Xp11.23: potential hotspot for neurogenetic disorders". Genomics. 79 (4): 560–72. doi:10.1006/geno.2002.6733. PMID 11944989.
- ^ Inoue A, Takahashi KP, Kimura M, Watanabe T, Morisawa S (August 1996). "Molecular cloning of a RNA binding protein, S1-1". Nucleic Acids Research. 24 (15): 2990–7. doi:10.1093/nar/24.15.2990. PMC 146028. PMID 8760884.
- ^ a b c d e f g Wang Y, Gogol-Döring A, Hu H, Fröhler S, Ma Y, Jens M, et al. (September 2013). "Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation". EMBO Molecular Medicine. 5 (9): 1431–42. doi:10.1002/emmm.201302663. PMC 3799496. PMID 24000153.
- ^ a b c d e f g h i j k l m Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J (December 2013). "RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation". Molecular Cell. 52 (5): 720–33. doi:10.1016/j.molcel.2013.11.010. hdl:10230/25288. PMID 24332178.
- ^ a b Inoue A, Yamamoto N, Kimura M, Nishio K, Yamane H, Nakajima K (March 2014). "RBM10 regulates alternative splicing". FEBS Letters. 588 (6): 942–7. doi:10.1016/j.febslet.2014.01.052. PMID 24530524. S2CID 10303057.
- ^ Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, et al. (February 2016). "Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing". Cell. 164 (4): 805–17. doi:10.1016/j.cell.2016.01.029. PMC 4882190. PMID 26871637.
- ^ a b Sutherland LC, Thibault P, Durand M, Lapointe E, Knee JM, Beauvais A, et al. (July 2017). "Splicing arrays reveal novel RBM10 targets, including SMN2 pre-mRNA". BMC Molecular Biology. 18 (1): 19. doi:10.1186/s12867-017-0096-x. PMC 5520337. PMID 28728573.
- ^ a b c d e Sun Y, Bao Y, Han W, Song F, Shen X, Zhao J, et al. (August 2017). "Autoregulation of RBM10 and cross-regulation of RBM10/RBM5 via alternative splicing-coupled nonsense-mediated decay". Nucleic Acids Research. 45 (14): 8524–8540. doi:10.1093/nar/gkx508. PMC 5737846. PMID 28586478.
- ^ a b Collins KM, Kainov YA, Christodolou E, Ray D, Morris Q, Hughes T, et al. (June 2017). "An RRM-ZnF RNA recognition module targets RBM10 to exonic sequences to promote exon exclusion". Nucleic Acids Research. 45 (11): 6761–6774. doi:10.1093/nar/gkx225. PMC 5499739. PMID 28379442.
- ^ a b c Loiselle JJ, Roy JG, Sutherland LC (2017). "RBM10 promotes transformation-associated processes in small cell lung cancer and is directly regulated by RBM5". PLOS ONE. 12 (6): e0180258. Bibcode:2017PLoSO..1280258L. doi:10.1371/journal.pone.0180258. PMC 5491171. PMID 28662214.
- ^ a b c d Johnston JJ, Teer JK, Cherukuri PF, Hansen NF, Loftus SK, Chong K, et al. (May 2010). "Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate". American Journal of Human Genetics. 86 (5): 743–8. doi:10.1016/j.ajhg.2010.04.007. PMC 2868995. PMID 20451169.
- ^ a b c Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. (September 2012). "Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing". Cell. 150 (6): 1107–20. doi:10.1016/j.cell.2012.08.029. PMC 3557932. PMID 22980975.
- ^ a b c Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P, et al. (April 2018). "Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types". Cell Reports. 23 (1): 282–296.e4. doi:10.1016/j.celrep.2018.01.088. PMC 5933844. PMID 29617667.
- ^ a b Cieply B, Carstens RP (2015). "Functional roles of alternative splicing factors in human disease". Wiley Interdisciplinary Reviews. RNA. 6 (3): 311–26. doi:10.1002/wrna.1276. PMC 4671264. PMID 25630614.
- ^ a b Coomer AO, Black F, Greystoke A, Munkley J, Elliott DJ (2019). "Alternative splicing in lung cancer". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1862 (11–12): 194388. doi:10.1016/j.bbagrm.2019.05.006. PMID 31152916. S2CID 173188639.
- ^ a b c Gorlin RJ, Cervenka J, Anderson RC, Sauk JJ, Bevis WD (February 1970). "Robin's syndrome. A probably X-linked recessive subvariety exhibiting persistence of left superior vena cava and atrial septal defect". American Journal of Diseases of Children. 119 (2): 176–8. doi:10.1001/archpedi.1970.02100050178020. PMID 5410571.
- ^ a b c d e Inoue A, Tsugawa K, Tokunaga K, Takahashi KP, Uni S, Kimura M, et al. (September 2008). "S1-1 nuclear domains: characterization and dynamics as a function of transcriptional activity". Biology of the Cell. 100 (9): 523–35. doi:10.1042/BC20070142. PMID 18315527. S2CID 9893063.
- ^ a b Zheng S, Damoiseaux R, Chen L, Black DL (June 2013). "A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing". Genome Research. 23 (6): 998–1007. doi:10.1101/gr.147546.112. PMC 3668367. PMID 23636947.
- ^ Lim J, Hao T, Shaw C, Patel AJ, Szabó G, Rual JF, et al. (May 2006). "A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration". Cell. 125 (4): 801–14. doi:10.1016/j.cell.2006.03.032. PMID 16713569. S2CID 13709685.
- ^ Mohan N, Kumar V, Kandala DT, Kartha CC, Laishram RS (September 2018). "A Splicing-Independent Function of RBM10 Controls Specific 3' UTR Processing to Regulate Cardiac Hypertrophy". Cell Reports. 24 (13): 3539–3553. doi:10.1016/j.celrep.2018.08.077. PMID 30257214.
- ^ a b c Mueller CF, Berger A, Zimmer S, Tiyerili V, Nickenig G (August 2009). "The heterogenous nuclear riboprotein S1-1 regulates AT1 receptor gene expression via transcriptional and posttranscriptional mechanisms". Archives of Biochemistry and Biophysics. 488 (1): 76–82. doi:10.1016/j.abb.2009.06.002. PMID 19508861.
- ^ Treiber T, Treiber N, Plessmann U, Harlander S, Daiß JL, Eichner N, et al. (April 2017). "A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis". Molecular Cell. 66 (2): 270–284.e13. doi:10.1016/j.molcel.2017.03.014. PMID 28431233.
- ^ a b c Jung JH, Lee H, Cao B, Liao P, Zeng SX, Lu H (January 2020). "RNA-binding motif protein 10 induces apoptosis and suppresses proliferation by activating p53". Oncogene. 39 (5): 1031–1040. doi:10.1038/s41388-019-1034-9. PMC 6994357. PMID 31591476.
- ^ Guan G, Li R, Tang W, Liu T, Su Z, Wang Y, et al. (March 2017). "Expression of RNA-binding motif 10 is associated with advanced tumor stage and malignant behaviors of lung adenocarcinoma cancer cells". Tumour Biology. 39 (3): 1010428317691740. doi:10.1177/1010428317691740. PMID 28347232. S2CID 206612545.
- ^ Kunimoto H, Inoue A, Kojima H, Yang J, Zhao H, Tsuruta D, Nakajima K (February 2020). "RBM10 regulates centriole duplication in HepG2 cells by ectopically assembling PLK4-STIL complexes in the nucleus". Genes to Cells. 25 (2): 100–110. doi:10.1111/gtc.12741. PMID 31820547. S2CID 209165475.
- ^ Pozzi B, Bragado L, Mammi P, Torti MF, Gaioli N, Gebhard LG, et al. (July 2020). "Dengue virus targets RBM10 deregulating host cell splicing and innate immune response". Nucleic Acids Research. 48 (12): 6824–6838. doi:10.1093/nar/gkaa340. PMC 7337517. PMID 32432721.
- ^ Salichs E, Ledda A, Mularoni L, Albà MM, de la Luna S (March 2009). "Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment". PLOS Genetics. 5 (3): e1000397. doi:10.1371/journal.pgen.1000397. PMC 2644819. PMID 19266028.
- ^ Goto Y, Kimura H (December 2009). "Inactive X chromosome-specific histone H3 modifications and CpG hypomethylation flank a chromatin boundary between an X-inactivated and an escape gene". Nucleic Acids Research. 37 (22): 7416–28. doi:10.1093/nar/gkp860. PMC 2794193. PMID 19843608.
- ^ Stes E, Laga M, Walton A, Samyn N, Timmerman E, De Smet I, et al. (June 2014). "A COFRADIC protocol to study protein ubiquitination". Journal of Proteome Research. 13 (6): 3107–13. doi:10.1021/pr4012443. PMID 24816145.
- ^ Akimov V, Barrio-Hernandez I, Hansen SV, Hallenborg P, Pedersen AK, Bekker-Jensen DB, et al. (July 2018). "UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites". Nature Structural & Molecular Biology. 25 (7): 631–640. doi:10.1038/s41594-018-0084-y. PMID 29967540. S2CID 49559977.
- ^ Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. (August 2009). "Lysine acetylation targets protein complexes and co-regulates major cellular functions". Science. 325 (5942): 834–40. Bibcode:2009Sci...325..834C. doi:10.1126/science.1175371. PMID 19608861. S2CID 206520776.
- ^ Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, et al. (January 2014). "Immunoaffinity enrichment and mass spectrometry analysis of protein methylation". Molecular & Cellular Proteomics. 13 (1): 372–87. doi:10.1074/mcp.O113.027870. PMC 3879628. PMID 24129315.
- ^ Cancer Genome Atlas Research Network (July 2014). "Comprehensive molecular profiling of lung adenocarcinoma". Nature. 511 (7511): 543–50. Bibcode:2014Natur.511..543T. doi:10.1038/nature13385. PMC 4231481. PMID 25079552.
- ^ Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao H, et al. (May 2016). "Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients". Cancer Cell. 29 (5): 711–722. doi:10.1016/j.ccell.2016.04.001. PMC 4864951. PMID 27165743.
- ^ Yin LL, Wen XM, Li M, Xu YM, Zhao XF, Li J, Wang XW (November 2018). "A gene mutation in RNA-binding protein 10 is associated with lung adenocarcinoma progression and poor prognosis". Oncology Letters. 16 (5): 6283–6292. doi:10.3892/ol.2018.9496. PMC 6202477. PMID 30405763.
- ^ Powis Z, Hart A, Cherny S, Petrik I, Palmaer E, Tang S, Jones C (2 June 2017). "Clinical diagnostic exome evaluation for an infant with a lethal disorder: genetic diagnosis of TARP syndrome and expansion of the phenotype in a patient with a newly reported RBM10 alteration". BMC Medical Genetics. 18 (1): 60. doi:10.1186/s12881-017-0426-3. PMC 5455125. PMID 28577551.
- ^ Gripp KW, Hopkins E, Johnston JJ, Krause C, Dobyns WB, Biesecker LG (October 2011). "Long-term survival in TARP syndrome and confirmation of RBM10 as the disease-causing gene". American Journal of Medical Genetics. Part A. 155A (10): 2516–20. doi:10.1002/ajmg.a.34190. PMC 3183328. PMID 21910224.
- ^ Niceta M, Barresi S, Pantaleoni F, Capolino R, Dentici ML, Ciolfi A, et al. (June 2019). "TARP syndrome: Long-term survival, anatomic patterns of congenital heart defects, differential diagnosis and pathogenetic considerations". European Journal of Medical Genetics. 62 (6): 103534. doi:10.1016/j.ejmg.2018.09.001. PMID 30189253.
- ^ Højland AT, Lolas I, Okkels H, Lautrup CK, Diness BR, Petersen MB, Nielsen IK (December 2018). "First reported adult patient with TARP syndrome: A case report". American Journal of Medical Genetics. Part A. 176 (12): 2915–2918. doi:10.1002/ajmg.a.40638. PMC 6587983. PMID 30462380.
- ^ Loiselle JJ, Sutherland LC (May 2018). "RBM10: Harmful or helpful-many factors to consider". Journal of Cellular Biochemistry. 119 (5): 3809–3818. doi:10.1002/jcb.26644. PMC 5901003. PMID 29274279.
- ^ Xia QY, Wang XT, Zhan XM, Tan X, Chen H, Liu Y, et al. (May 2017). "Xp11 Translocation Renal Cell Carcinomas (RCCs) With RBM10-TFE3 Gene Fusion Demonstrating Melanotic Features and Overlapping Morphology With t(6;11) RCC: Interest and Diagnostic Pitfall in Detecting a Paracentric Inversion of TFE3". The American Journal of Surgical Pathology. 41 (5): 663–676. doi:10.1097/PAS.0000000000000837. PMID 28288037. S2CID 205918230.
- ^ Argani P, Zhang L, Reuter VE, Tickoo SK, Antonescu CR (May 2017). "RBM10-TFE3 Renal Cell Carcinoma: A Potential Diagnostic Pitfall Due to Cryptic Intrachromosomal Xp11.2 Inversion Resulting in False-negative TFE3 FISH". The American Journal of Surgical Pathology. 41 (5): 655–662. doi:10.1097/PAS.0000000000000835. PMC 5391276. PMID 28296677.
- ^ Kato I, Furuya M, Baba M, Kameda Y, Yasuda M, Nishimoto K, et al. (August 2019). "RBM10-TFE3 renal cell carcinoma characterised by paracentric inversion with consistent closely split signals in break-apart fluorescence in-situ hybridisation: study of 10 cases and a literature review". Histopathology. 75 (2): 254–265. doi:10.1111/his.13866. PMID 30908700. S2CID 85516169.
- ^ a b Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. (April 2015). "Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets". Nature Communications. 6: 6744. Bibcode:2015NatCo...6.6744W. doi:10.1038/ncomms7744. PMC 4403382. PMID 25855536.
- ^ Furukawa T, Kuboki Y, Tanji E, Yoshida S, Hatori T, Yamamoto M, et al. (2011). "Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas". Scientific Reports. 1: 161. Bibcode:2011NatSR...1E.161F. doi:10.1038/srep00161. PMC 3240977. PMID 22355676.
- ^ Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. (April 2016). "Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma". Cell Reports. 15 (4): 857–865. doi:10.1016/j.celrep.2016.03.075. PMC 4850357. PMID 27149842.
- ^ Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. (January 2014). "Discovery and saturation analysis of cancer genes across 21 tumour types". Nature. 505 (7484): 495–501. Bibcode:2014Natur.505..495L. doi:10.1038/nature12912. PMC 4048962. PMID 24390350.
- ^ Ibrahimpasic T, Xu B, Landa I, Dogan S, Middha S, Seshan V, et al. (October 2017). "RBM10 as Novel Thyroid Cancer Genes Associated with Tumor Virulence". Clinical Cancer Research. 23 (19): 5970–5980. doi:10.1158/1078-0432.CCR-17-1183. PMC 5626586. PMID 28634282.
- ^ Antonello ZA, Hsu N, Bhasin M, Roti G, Joshi M, Van Hummelen P, et al. (October 2017). "V600E". Oncotarget. 8 (49): 84743–84760. doi:10.18632/oncotarget.21262. PMC 5689570. PMID 29156680.
- ^ Ibrahimpasic T, Ghossein R, Shah JP, Ganly I (March 2019). "Poorly Differentiated Carcinoma of the Thyroid Gland: Current Status and Future Prospects". Thyroid. 29 (3): 311–321. doi:10.1089/thy.2018.0509. PMC 6437626. PMID 30747050.
- ^ a b Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. (August 2010). "Diverse somatic mutation patterns and pathway alterations in human cancers". Nature. 466 (7308): 869–73. Bibcode:2010Natur.466..869K. doi:10.1038/nature09208. PMC 3026267. PMID 20668451.
- ^ Tian W, Hu W, Shi X, Liu P, Ma X, Zhao W, et al. (April 2020). "Comprehensive genomic profile of cholangiocarcinomas in China". Oncology Letters. 19 (4): 3101–3110. doi:10.3892/ol.2020.11429. PMC 7074170. PMID 32256810.
- ^ Schwab ME, Song H, Mattis A, Phelps A, Vu LT, Huang FW, Nijagal A (March 2020). "De novo somatic mutations and KRAS amplification are associated with cholangiocarcinoma in a patient with a history of choledochal cyst". Journal of Pediatric Surgery. 55 (12): 2657–2661. doi:10.1016/j.jpedsurg.2020.03.008. PMC 7942710. PMID 32295706.
- ^ Juratli TA, McCabe D, Nayyar N, Williams EA, Silverman IM, Tummala SS, et al. (November 2018). "DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome". Acta Neuropathologica. 136 (5): 779–792. doi:10.1007/s00401-018-1899-7. PMID 30123936. S2CID 52039057.
- ^ Majd NK, Metrus NR, Santos-Pinheiro F, Trevino CR, Fuller GN, Huse JT, et al. (February 2019). "RBM10 truncation in astroblastoma in a patient with history of mandibular ameloblastoma: A case report". Cancer Genetics. 231–232: 41–45. doi:10.1016/j.cancergen.2019.01.001. PMID 30803556. S2CID 73477150.
- ^ Misquitta-Ali CM, Cheng E, O'Hanlon D, Liu N, McGlade CJ, Tsao MS, Blencowe BJ (January 2011). "Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer". Molecular and Cellular Biology. 31 (1): 138–50. doi:10.1128/MCB.00709-10. PMC 3019846. PMID 21041478.
- ^ a b Hernández J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcárcel J (2016). "Tumor suppressor properties of the splicing regulatory factor RBM10". RNA Biology. 13 (4): 466–72. doi:10.1080/15476286.2016.1144004. PMC 4841610. PMID 26853560.
- ^ a b c Zhao J, Sun Y, Huang Y, Song F, Huang Z, Bao Y, et al. (January 2017). "Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing". Scientific Reports. 7: 40488. Bibcode:2017NatSR...740488Z. doi:10.1038/srep40488. PMC 5238425. PMID 28091594.
- ^ a b Han LP, Wang CP, Han SL (October 2018). "Overexpression of RBM10 induces osteosarcoma cell apoptosis and inhibits cell proliferation and migration". Médecine/Sciences. 34 Focus issue F1: 81–86. doi:10.1051/medsci/201834f114. PMID 30403180.
- ^ Jin X, Di X, Wang R, Ma H, Tian C, Zhao M, et al. (June 2019). "RBM10 inhibits cell proliferation of lung adenocarcinoma via RAP1/AKT/CREB signalling pathway". Journal of Cellular and Molecular Medicine. 23 (6): 3897–3904. doi:10.1111/jcmm.14263. PMC 6533519. PMID 30955253.
- ^ Sutherland LC, Rintala-Maki ND, White RD, Morin CD (January 2005). "RNA binding motif (RBM) proteins: a novel family of apoptosis modulators?". Journal of Cellular Biochemistry. 94 (1): 5–24. doi:10.1002/jcb.20204. PMID 15514923. S2CID 26344717.
- ^ Wang K, Bacon ML, Tessier JJ, Rintala-Maki ND, Tang V, Sutherland LC (2012). "RBM10 Modulates Apoptosis and Influences TNF-α Gene Expression". Journal of Cell Death. 5: 1–19. doi:10.4137/JCD.S9073. PMC 4583097. PMID 26446321.
- ^ Rodor J, FitzPatrick DR, Eyras E, Cáceres JF (January 2017). "The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development". RNA Biology. 14 (1): 45–57. doi:10.1080/15476286.2016.1247148. PMC 5270529. PMID 27763814.
- ^ Sun X, Jia M, Sun W, Feng L, Gu C, Wu T (February 2019). "Functional role of RBM10 in lung adenocarcinoma proliferation". International Journal of Oncology. 54 (2): 467–478. doi:10.3892/ijo.2018.4643. PMC 6317669. PMID 30483773.
- ^ Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. (November 2017). "Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer". Nature. 551 (7681): 512–516. Bibcode:2017Natur.551..512B. doi:10.1038/nature24462. PMC 6145146. PMID 29132146.
- ^ Siegel RL, Miller KD, Jemal A (January 2018). "Cancer statistics, 2018". CA: A Cancer Journal for Clinicians. 68 (1): 7–30. doi:10.3322/caac.21442. PMID 29313949.
- ^ Mourtada-Maarabouni M, Williams GT (July 2002). "RBM5/LUCA-15--tumour suppression by control of apoptosis and the cell cycle?". TheScientificWorldJournal. 2: 1885–90. doi:10.1100/tsw.2002.859. PMC 6009235. PMID 12920317.
- ^ Oh JJ, Razfar A, Delgado I, Reed RA, Malkina A, Boctor B, Slamon DJ (April 2006). "3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis". Cancer Research. 66 (7): 3419–27. doi:10.1158/0008-5472.CAN-05-1667. PMID 16585163.
- ^ Fushimi K, Ray P, Kar A, Wang L, Sutherland LC, Wu JY (October 2008). "Up-regulation of the proapoptotic caspase 2 splicing isoform by a candidate tumor suppressor, RBM5". Proceedings of the National Academy of Sciences of the United States of America. 105 (41): 15708–13. Bibcode:2008PNAS..10515708F. doi:10.1073/pnas.0805569105. PMC 2572934. PMID 18840686.
- ^ Bonnal S, Martínez C, Förch P, Bachi A, Wilm M, Valcárcel J (October 2008). "RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition". Molecular Cell. 32 (1): 81–95. doi:10.1016/j.molcel.2008.08.008. PMID 18851835.
- ^ Sutherland LC, Wang K, Robinson AG (March 2010). "RBM5 as a putative tumor suppressor gene for lung cancer". Journal of Thoracic Oncology. 5 (3): 294–8. doi:10.1097/JTO.0b013e3181c6e330. PMID 20186023.
- ^ Jamsai D, Watkins DN, O'Connor AE, Merriner DJ, Gursoy S, Bird AD, et al. (November 2017). "In vivo evidence that RBM5 is a tumour suppressor in the lung". Scientific Reports. 7 (1): 16323. Bibcode:2017NatSR...716323J. doi:10.1038/s41598-017-15874-9. PMC 5701194. PMID 29176597.
- ^ Wang Q, Wang F, Zhong W, Ling H, Wang J, Cui J, et al. (May 2019). "RNA-binding protein RBM6 as a tumor suppressor gene represses the growth and progression in laryngocarcinoma". Gene. 697: 26–34. doi:10.1016/j.gene.2019.02.025. PMID 30772516. S2CID 73456531.
- ^ Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, et al. (July 2006). "Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions". Molecular and Cellular Biology. 26 (14): 5528–43. doi:10.1128/MCB.00582-06. PMC 1592722. PMID 16809785.
- ^ Papasaikas P, Tejedor JR, Vigevani L, Valcárcel J (January 2015). "Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery". Molecular Cell. 57 (1): 7–22. doi:10.1016/j.molcel.2014.10.030. hdl:10230/45908. PMID 25482510. S2CID 11133534.
- ^ Ule J, Blencowe BJ (October 2019). "Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution". Molecular Cell. 76 (2): 329–345. doi:10.1016/j.molcel.2019.09.017. PMID 31626751.
Further reading[edit]
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Hartley JL, Temple GF, Brasch MA (November 2000). "DNA cloning using in vitro site-specific recombination". Genome Research. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
- Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, et al. (March 2001). "Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs". Genome Research. 11 (3): 422–35. doi:10.1101/gr.GR1547R. PMC 311072. PMID 11230166.
- Li J, Hawkins IC, Harvey CD, Jennings JL, Link AJ, Patton JG (November 2003). "Regulation of alternative splicing by SRrp86 and its interacting proteins". Molecular and Cellular Biology. 23 (21): 7437–47. doi:10.1128/MCB.23.21.7437-7447.2003. PMC 207616. PMID 14559993.
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villén J, Li J, et al. (August 2004). "Large-scale characterization of HeLa cell nuclear phosphoproteins". Proceedings of the National Academy of Sciences of the United States of America. 101 (33): 12130–5. Bibcode:2004PNAS..10112130B. doi:10.1073/pnas.0404720101. PMC 514446. PMID 15302935.
- Ballif BA, Villén J, Beausoleil SA, Schwartz D, Gygi SP (November 2004). "Phosphoproteomic analysis of the developing mouse brain". Molecular & Cellular Proteomics. 3 (11): 1093–101. doi:10.1074/mcp.M400085-MCP200. PMID 15345747.
- Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, et al. (October 2004). "From ORFeome to biology: a functional genomics pipeline". Genome Research. 14 (10B): 2136–44. doi:10.1101/gr.2576704. PMC 528930. PMID 15489336.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Mehrle A, Rosenfelder H, Schupp I, del Val C, Arlt D, Hahne F, et al. (January 2006). "The LIFEdb database in 2006". Nucleic Acids Research. 34 (Database issue): D415-8. doi:10.1093/nar/gkj139. PMC 1347501. PMID 16381901.
- Martínez-Arribas F, Agudo D, Pollán M, Gómez-Esquer F, Díaz-Gil G, Lucas R, Schneider J (April 2006). "Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer". Journal of Cellular Biochemistry. 97 (6): 1275–82. doi:10.1002/jcb.20725. PMID 16552754. S2CID 9804734.
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (November 2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983. S2CID 7827573.