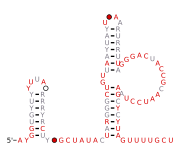RT RNA motifs
Appearance
This article has multiple issues. Please help improve it or discuss these issues on the talk page. (Learn how and when to remove these messages)
|
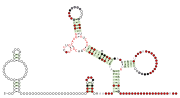
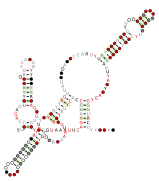
RT RNA motifs refers to conserved RNA motifs discovered by bioinformatics and that are usually or always located nearby to genes predicted to encode reverse transcriptase (RT genes). Known RNAs located nearby to RT genes include self-splicing introns, retrons and diversity-generating retroelements (DGR), and RT RNA motifs could function as part of such elements.
Nineteen RT RNA motifs found and named RT-1 through RT-19.[1] The RT-10 RNA motif occurs in a known DGR in Bordetella phage BPP-1. However, the function of these RNAs and their role in the DGR remains unknown. Most RT RNA motifs to date appear too small to be likely to function as self-splicing RNAs.
References
[edit]- ^ Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR (October 2017). "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. doi:10.1093/nar/gkx699. PMC 5737381. PMID 28977401.