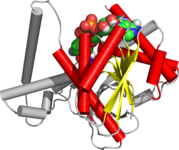
Rossmann fold
This article is missing information about more non-nucleotide functions. (February 2021) |
| Rossmann-like alpha/beta/alpha sandwich fold | |
|---|---|
 NAD/NADP binding rossmann fold domains. The picture depicts the beta-alpha folding in alcohol dehydrogenase. | |
| Identifiers | |
| Symbol | Rossmann-like_a/b/a_fold |
| Pfam clan | CL0039 |
| InterPro | IPR014729 |
The Rossmann fold is a tertiary fold found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. This fold is composed of alternating beta strands and alpha helical segments where the beta strands are hydrogen bonded to each other forming an extended beta sheet and the alpha helices surround both faces of the sheet to produce a three-layered sandwich. The classical Rossmann fold contains six beta strands whereas Rossmann-like folds, sometimes referred to as Rossmannoid folds, contain only five strands. The initial beta-alpha-beta (bab) fold is the most conserved segment of the Rossmann fold.[1] The motif is named after Michael Rossmann who first noticed this structural motif in the enzyme lactate dehydrogenase in 1970 and who later observed that this was a frequently occurring motif in nucleotide binding proteins.[2]
Rossmann and Rossmannoid fold proteins are extremely common. They make up 20% of proteins with known structures in the Protein Data Bank, and are found in more than 38% of KEGG metabolic pathways.[3] The fold is extremely versatile in that it can accommodate a wide range of ligands. They can function as metabolic enzymes, DNA/RNA binding, and regulatory proteins in addition to the traditional role.[4]
History[edit]
The Rossmann fold was first described by Dr. Michael Rossmann and coworkers in 1974.[5] He was the first to deduce the structure of lactate dehydrogenase and characterized the structural motif within this enzyme which would later be called the Rossmann fold. It was subsequently found that most dehydrogenases that utilize NAD or NADP contain this same structurally conserved Rossmann fold motif.[5][6]
In 1989, Israel Hanukoglu from the Weizmann Institute of Science discovered that the consensus sequence for NADP+ binding site in some enzymes that utilize NADP+ differs from the NAD+ binding motif.[7] This discovery was used to re-engineer coenzyme specificities of enzymes.[8]
Structure[edit]

The Rossmann fold is composed of six parallel beta strands that form an extended beta sheet. The first three strands are connected by α- helices resulting in a beta-alpha-beta-alpha-beta structure. This pattern is duplicated once to produce an inverted tandem repeat containing six strands. Overall, the strands are arranged in the order of 321456 (1 = N-terminal, 6 = C-terminal).[9] Five stranded Rossmann-like folds are arranged in the order 32145.[10] The overall tertiary structure of the fold resembles a three-layered sandwich wherein the filling is composed of an extended beta sheet and the two slices of bread are formed by the connecting parallel alpha-helices.[1]
One of the features of the Rossmann fold is its co-factor binding specificity. Through the analysis of four NADH-binding enzymes, it was found that in all four enzymes the nucleotide co-factor entailed the same conformation and orientation with respect to the polypeptide chain.[1]
The fold may contain additional strands joined by short helices or coils.[1] The most conserved segment of Rossmann folds is the first beta-alpha-beta segment. Phosphate-binding loop is located between the first beta-strand and alpha-helix. On the tip of the second beta-strand, there is a conserved aspartate residue that is involved in ribose binding.[11] Since this segment is in contact with the ADP portion of dinucleotides such as FAD, NAD and NADP it is also called as an "ADP-binding beta-beta fold.
Function[edit]
The function of the Rossmann fold in enzymes is to bind nucleotide cofactors. It also often contributes to substrate binding.
Metabolic enzymes normally have one specific function, and in the case of UDP-glucose 6-dehydrogenase, the primary function is to catalyze the two step NAD(+)-dependent oxidation of UDP-glucose into UDP-glucuronic acid.[12] The N- and C-terminal domains of UgdG share structural features with ancient mitochondrial ribonucleases named MAR. MARs are present in lower eukaryotic microorganisms, have a Rossmannoid-fold and belong to the isochorismatase superfamily. This observation reinforces that the Rossmann structural motifs found in NAD(+)-dependent dehydrogenases can have a dual function working as a nucleotide cofactor binding domain and as a ribonuclease.
Evolution[edit]
Rossman and Rossmannoids[edit]
The evolutionary relationship between the Rossmann fold and Rossmann-like folds is unclear. These folds are referred to as Rossmannoids. It has been hypothesized that all these folds, including a Rossmann fold originated from a single common ancestral fold, that had nucleotide binding capabilities, in addition to non-specific catalytic activity.[5]
However, an analysis of the PDB finds evidence of convergent evolution[3] with 156 separate H-groups of demonstrable homology, from which 123 X-groups of probable homology can be found. The groups have been integrated into ECOD.[4]
Conventional Rossman group[edit]
Phylogenetic analysis of the NADP binding enzyme adrenodoxin reductase revealed that from prokaryotes, through metazoa and up to primates the sequence motif difference from that of most FAD and NAD-binding sites is strictly conserved.[13]
In many articles and textbooks, a Rossmann fold is defined as a strict repeated series of βαβ structure. Yet, comprehensive examination of the Rossmann folds in many NAD(P) and FAD binding sites revealed that only the first βα structure is strictly conserved. In some enzymes, there may be many loops and several helices (i.e., not a single helix) between the beta strands that form the beta-sheet.[1] These enzymes have a common origin indicated by conserved sequence and structural features, according to Hanukoglu.[13]
The result by Hanukoglu (2017) is corroborated by Medvedev et al. (2020), in the form of an ECOD "H-group" called "Rossmann-related". Even within this group, ECOD describes a wide range of non-nucleotide activities.[4]
References[edit]
- ^ a b c d e Hanukoglu I (2015). "Proteopedia: Rossmann fold: A beta-alpha-beta fold at dinucleotide binding sites". Biochemistry and Molecular Biology Education. 43 (3): 206–9. doi:10.1002/bmb.20849. PMID 25704928.
- ^ Cox MM, Nelson DL (2013). Lehninger Principles of Biochemistry (6th ed.). New York: W.H. Freeman. ISBN 978-1-4292-3414-6.
- ^ a b Medvedev KE, Kinch LN, Schaeffer RD, Grishin NV (December 2019). "Functional analysis of Rossmann-like domains reveals convergent evolution of topology and reaction pathways". PLOS Computational Biology. 15 (12): e1007569. Bibcode:2019PLSCB..15E7569M. doi:10.1371/journal.pcbi.1007569. PMC 6957218. PMID 31869345.
- ^ a b c Medvedev KE, Kinch LN, Dustin Schaeffer R, Pei J, Grishin NV (February 2021). "A Fifth of the Protein World: Rossmann-like Proteins as an Evolutionarily Successful Structural unit". Journal of Molecular Biology. 433 (4): 166788. doi:10.1016/j.jmb.2020.166788. PMC 7870570. PMID 33387532.
Medvedev KE, et al. "Rossmann-fold project". Grishin Lab. UT Southwestern Medical Center. - ^ a b c Kessel A (2010). Introduction to Proteins: Structure, Function, and Motion. Florida: CRC Press. p. 143. ISBN 978-1-4398-1071-2.
- ^ Rao ST, Rossmann MG (May 1973). "Comparison of super-secondary structures in proteins". Journal of Molecular Biology. 76 (2): 241–56. doi:10.1016/0022-2836(73)90388-4. PMID 4737475.
- ^ Hanukoglu I, Gutfinger T (March 1989). "cDNA sequence of adrenodoxin reductase. Identification of NADP-binding sites in oxidoreductases". European Journal of Biochemistry. 180 (2): 479–84. doi:10.1111/j.1432-1033.1989.tb14671.x. PMID 2924777.
- ^ Scrutton NS, Berry A, Perham RN (January 1990). "Redesign of the coenzyme specificity of a dehydrogenase by protein engineering". Nature. 343 (6253): 38–43. Bibcode:1990Natur.343...38S. doi:10.1038/343038a0. PMID 2296288. S2CID 1580419.
- ^ "NAD(P)-binding Rossmann-fold domains". SCOP: Structural Classification of Proteins. Archived from the original on 2018-11-21. Retrieved 2017-12-17.
- ^ "Nucleotide-binding domain". SCOP: Structural Classification of Proteins. Archived from the original on 2018-12-07. Retrieved 2017-12-17.
- ^ Longo LM, Jabłońska J, Vyas P, Kanade M, Kolodny R, Ben-Tal N, Tawfik DS (December 2020). Deane CM, Boudker O (eds.). "On the emergence of P-Loop NTPase and Rossmann enzymes from a Beta-Alpha-Beta ancestral fragment". eLife. 9: e64415. doi:10.7554/eLife.64415. PMC 7758060. PMID 33295875.
- ^ Bhattacharyya M, Upadhyay R, Vishveshwara S (2012). "Interaction signatures stabilizing the NAD(P)-binding Rossmann fold: a structure network approach". PLOS ONE. 7 (12): e51676. Bibcode:2012PLoSO...751676B. doi:10.1371/journal.pone.0051676. PMC 3524241. PMID 23284738.
- ^ a b Hanukoglu I (2017). "Conservation of the Enzyme-Coenzyme Interfaces in FAD and NADP Binding Adrenodoxin Reductase-A Ubiquitous Enzyme". Journal of Molecular Evolution. 85 (5): 205–218. Bibcode:2017JMolE..85..205H. doi:10.1007/s00239-017-9821-9. PMID 29177972. S2CID 7120148.