T-even bacteriophages
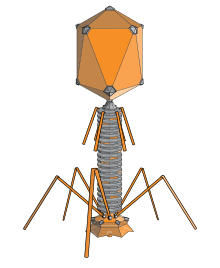
T-even phages, also known as the E. coli phages, are a group of double-stranded bacteriophages from the family Myoviridae. Bacteriophage means to "eat bacteria", and phages are well known for being obligate intracellular parasites that reproduce within the host cell and are released when the host is destroyed by lysis. Containing about 160 genes, these virulent viruses are among the largest, most complex viruses that are known and one of the best studied model organisms. They have played a key role in the development of virology and molecular biology.[1][2]
Discovery
Bacteriophages were first discovered by the English scientist Frederick Twort in 1915 and Félix d'Hérelle in 1917. In the late 1930s, T.L. Rakieten proposed either a mixture of raw sewerage or a lysate from E.coli infected with raw sewerage to the two researchers Milislav Demerac and Ugo Fano. These two researchers isolated T3, T4,T5, and T6 from E.coli. Also, in 1932, the researcher J.Bronfenbrenner had studied and worked on the T2 phage, at which the T2 phage was isolated from the virus.[3] This isolation was made from a fecal material rather than from sewerage. At any rate, Delbruck was involved in the discovery of the T even phages. His part was naming the bacteriophages into Type 1(T1), Type 2 (T2),Type 3 (T3), and etc.
Structure
Phages have multiple factors contributing to their structure. It consists of the head, collar, helical sheath, the core or tube, hexagonal base plate, tail fibers (not all) and finally tail pins. The head’s job is to enfold or surround nucleic acids. The tail fiber’s help in attaching the phage to a bacterial cell. The tail acts as a duct through which the nucleic acid goes through during an infection. The collar helps support the head (to stay in place). Bacteriophages in general (including T-even bacteriophages) contain a head structure, which can vary in size and shape. The head enfolds nucleic acid and acts as the protective covering.[4] Certain phages have tails attached to the phage head. The tail is a hollow duct through which the nucleic acid passes during infection. T-even Bacteriophages have genomes that code for phage-specific DNA replication, recombination, and DNA repair functions. Also, they offer well branded genes and proteins. Similar to all viruses, they depend on many of their hosts important makeups and roles or functions (transcription and translation, membranes, breakdown of energy etc.), for their reproduction.
Unique Features of the T-even Phages
Dating back to the 1940s till date, T-even phages are considered the best studied model organisms. Model organisms are usually required to be simple with as few as five genes. Yet, T-even phages are in fact among the largest and highest complexity virus, in which these phages genetic information is made up of around 160 genes. Coincident with their complexity, T-even viruses were found to have an unimaginable feature of no other, the presence of the unusual base hydroxymethylcytosine (HMC) in place of the nucleic acid base cytosine. In addition to this, the HMC residues on the T-even phage are glucosylated in a specific pattern. This unique feature allowed the formation of new enzymes that never existed in T-even infected cells or any other cell and modifying T-even phage DNA provided a basic underlying advancement in viral and molecular levels. Other unique features of the T-even virus is its regulated gene expression.[5] These unique features and other features gave significance of the T-even phages, this includes transduction which is responsible for transfer of drug resistant features, lysogenic conversion is responsible for acquisition of new characteristics such as the formation of new enzymes, random insertion into bacterial chromosome can induce insertional mutation, epidemiological typing of bacteria (phage typing), phages are used extensively in genetic engineering where they serve as cloning vectors. Libraries of genes and monoclonal antibodies are maintained in phages. In addition to all this they are responsible for natural removal of bacteria from water bodies.[6]
Life Cycle
This section needs additional citations for verification. (May 2016) |

Adsorption and Penetration
Just like all other viruses, T-even phages don't just randomely attach to the surface of their host; instead they "search" and bind to receptors, specific protein structures, found on the surface of the host. These receptors vary with the phage; teichoic acid, cell wall proteins and lipopolysaccharides, flagella, and pilis all can serve as receptors for the phage to bind to. In order for the T-even phage to infect its host and begin its life cycle it must enter the first process of infection, adsorption of the phage to the bacterial cell. Adsorption is a value characteristic of phage-host pair and the adsorption of the phage on host cell surface is illustrated as a 2-stage process: reverrsible and irreversible. It involves the phages tail structure that begins when the phages tail fibers helps bind the phage to the appropriate receptor of its host. This process is reversible. One or more of the components of the base plate mediates irreversible process of binding of the phage to a bacterium.

Penetration is also a value characteristic of phage-host infection that involves the injection of the phages genetic material inside the bacterium. Penetration of nucleic acid takes place after the irreversible adsorption phase. Mechanisms involving penetration of the phages nucleic acid are specific for each phage. This penetration mechanism can involve electrochemical membrane potential, ATP molecules, enzymatic splitting of peptidoglycan layer, or all three of these factor can be vital for the penetration of the nucleic acid inside the bacterial cell. Studies have been done on the T2 bacteriophage (T4-like phage) mechanism of penetration and it has shown that the phages tail does not penetrate inside the bacterial cell wall and penetration of this phage involves electrochemical membrane potential on the inner membrane.[7][8]
Lytic Cycle

Virulent bacteriophages multiply in their bacterial host immediately after entry. After the number of progeny phages reach a certain amount, they cause the host to lyse or break down, therefore they would be released and infect new host cells.[9] The process of host lyses and release is called the lytic cycle. Lytic cycle is a cycle of viral reproduction that involves the destruction of the infected cell and its membrane. This cycle involves a virus that overtakes the host cell and its machinery to reproduce. Therefore, the virus must go through 5 stages inorder to reproduce and infect the host cell. Such stages are Attachment, Penetration, Biosynthesis, Maturation, and then Release.
Attachment
Attachment occurs when the bacteria and the phage particles come in contact. This actually happens when the attachment site found on the viral surface attaches to the complementary receptor site on the bacteria. Attachment is supported by weak chemical bonds between the attachment and the receptor site. Also, the fibers at the end of the T even bacteriophage tail plays a role in the attachment site. Therefore, there must be specific recetor attachment as the receptor determines the host preference.
Penetration
This stage involves for the virus to penetrate itself in the host cell. It does so by injecting its DNA into the cell. The process of penetration happens when the bacteriophage's tail secretes the enzyme lysozyme, which breaks down the bacterial cell wall into segments. Therefore, the break down of the cell by the enzyme lysozyme forms a hole on the host cell;so, the bacteriophage inserts the DNA (genetic material) within the cell as the capsid is kept outside the cell.
Biosynthesis
After the bacterial DNA is inserted into the cell, the process of biosynthesis is initiated. Biosynthesis involves the utilization of the host cells' nucleotides and enzymes to make copies of the phage DNA. The mRNA in the cytoplasm are transcribed from the phage DNA being involved in the biosynthesis of phage enzymes and capsids proteins. Therefore, the ribosomes, enzymes, and amino acids of the host are involved in translation.
Maturation
Maturation is submitted by the assembly of bacteriophage DNA and capsids into virions. Late proteins are important in assembly. Assembly is complicated but varies in bacteriophage stages, some are assembled in nucleus and some are assembled in cytoplasm, and may be seen as paracrystalline structures in cell.The tails and heads of the T even bacteriophage are assembled from protein subunits, and the head is packed with DNA and attached to the tail.
Release
The final step in viral reproduction and multiplication is determined by the release of virions from the host cell. The release of the virions occurs after the breakage of the bacterial plasma membrane. Nonenveloped viruses lyse the host cell which is characterized by viral proteins attacking the peptidoglycan or membrane. The lysis of the bacteria occurs when the capsids inside the cell release the enzyme lysozyme which break down the cell wall. The released bacteriophages infect other cells, and the viral multiplication cycle is repeated within those cells.
Lysogenic Cycle
This section needs additional citations for verification. (May 2016) |

Lysogenic phages multiply in one of two ways; either by entering an inactive or latent state or by multiplying through the lytic phase. Through a process known as lysogeny, the phage DNA replicates with the replication of the host chromosome by assimilating into the host chromosome itself. It is then passed on to its daughter cells; this is why it usually isn’t recognized by the host. A process known as lysogenic or phage conversion changes the properties of the bacteria cell; this is possible because the prophage itself contains genes that can present new properties into the bacteria cell or host cell. These bacteria are considered lysogenized. Lysogenized bacteria are resistant to superinfection by same or related phages. This is known as superinfection immunity.[10]
T even phage Genome
T even bacteriophages are genetically related. The genetic diversity of bacteriophage is extraordinary and phenomenal. The genetic elements of a bacteriophage is transferred from one bacterium to another through the process of transduction. The process of transduction can be specialized or generalized. The generalized transduction is found within the lytic cycle where units of the bacterial DNA are packaged in a phage capsid.[11] Therefore, when these type of phages infect other bacterial cells, the bacterial DNA is inserted inside. Previously mentioned, bacterial genes can be transferred in generalized transduction. Whereas, in specialized transduction, only the genes that are adjacent to the prophage are transferred. Bacteriophage genomes can be highly mosaic at which the phage is made up of individual modules which may be found in other phage species in different arrangements. Based on correlative sequence similarity regulatory patterns, genomic organization, and virion structureof their essential genes, other phages from various parts of the world belong to this family.
See also
References
- ^ Norkin, Leonard C. (2010). Virology, Molecular Biology and Pathogenesis. Washington: American Society for Microbiology. p. 725. ISBN 978-1-55581-453-3.
- ^ Prescott, Harley, and Klein (2008). Microbiology (seventh ed.). McGraw Hill. p. 1078. ISBN 978-007-126727-4.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Willey, Joanne. Prescott's Microbiology (seventh ed.). McGraw-Hill.
- ^ Prescott,Harley, ,and Klein (2008). Microbiology (seventh ed.). McGraw-Hill. ISBN 978-007-126727-4.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Norkin, Leonard C. (2010). Virology, Molecular Biology and Pathogenesis. Washington: American Society for Microbiology. p. 725. ISBN 978-1-55581-453-3.
- ^ Rao, Srdhar. "Bacteriophages" (PDF). microrao.
- ^ Norkin, Leonard C. (2010). Virology, Molecular Biology and Pathogenesis. Washington: American Society for Microbiology. p. 31. ISBN 978-1-55581-453-3.
- ^ Prescott, Harley, and Klein (2008). Microbiology (seventh ed.). McGraw Hill. p. 427. ISBN 978-007-126727-4.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Sherwood, Linda (2011). Prescott's Microbiology (eighth ed.). McGraw-Hill.
- ^ Willey, Joanne (2011). Prescott's Microbiology (eighth ed.). McGraw-Hill.
- ^ Leonard C., Norkin (2010). Virology, Molecular Biology and Pathogenesis. Washington: American Society for Microbiology.
