Tractography

In neuroscience, tractography is a procedure to demonstrate the neural tracts.[1] It uses special techniques of magnetic resonance imaging (MRI), and computer-based image analysis. The results are presented in two- and three-dimensional images.
In addition to the long tracts that connect the brain to the rest of the body, there is a complicated 3D network formed by short connections among different cortical and subcortical regions. The existence of these bundles has been revealed by histochemistry and biological techniques on post-mortem specimens. Brain tracts are not identifiable by direct exam, CT, or MRI scans. This difficulty explains the paucity of their description in neuroanatomy atlases and the poor understanding of their functions.
The MRI sequences used look at the symmetry of brain water diffusion. Bundles of fiber tracts make the water diffuse asymmetrically in a tensor, the major axis parallel to the direction of the fibers. The asymmetry here is called anisotropy. There is a direct relationship between the number of fibers and the degree of anisotropy.
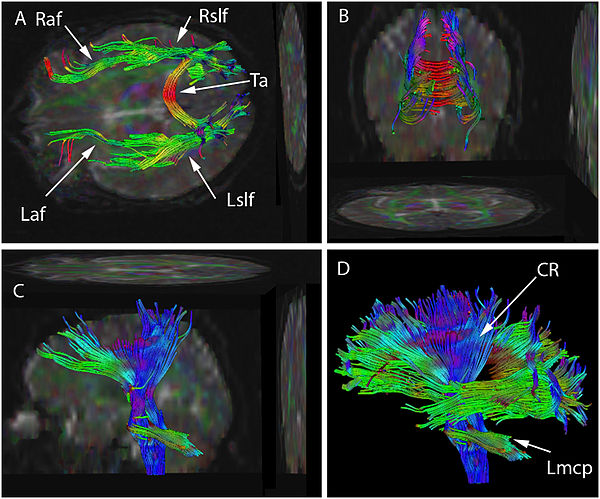
Figure legend: Diffusion tensor imaging (DTI) data has been used to seed various tractographic assessments of this patient's brain. These are seen in superior (A), posterior (B), and lateral views (C&D). The seeds have been used to develop arcuate and superior longitudinal fasciculi in (A) and (B), for brainstem, and corona radiata in (C), and as combined data sets in (D). Some of the two dimensional projections of the tractographic result are also shown. The data set may be rotated continuously into various planes to better appreciate the structure. Color has been assigned based on the dominant direction of the fibers. There is asymmetry in the tractographic fiber volume between the right and left arcuate fasciculus (Raf & Laf) (smaller on the left) and between the right and left superior longitudinal fasciculus (Rslf & Lslf) (smaller on the right). Also seen are Tapetum (Ta), Left corona radiata (Lcr) and Left middle cerebellar peduncle (Lmcp).
MRI technique

Tractography is performed using Diffusion Tensor Imaging, an MR technique which is sensitive to the diffusion of water in the body, and can be used to reveal its 3D shape. Free diffusion occurs equally in all directions. This is termed "isotropic" diffusion. If the water diffuses in a medium with barriers, the diffusion will be uneven, which is termed "anisotropic" diffusion. In such a case, the relative mobility of the molecules from the origin has a shape different from a sphere. This shape is often modeled as an ellipsoid, and the technique is then called Diffusion Tensor Imaging. Barriers can be many things—cell membranes, axons, myelin, etc.; but in white matter the principal barrier is the myelin sheath of axons. Bundles of axons provide a barrier to perpendicular diffusion and a path for parallel diffusion along the orientation of the fibers.
Anisotropic diffusion is expected to be increased in areas of high mature axonal order. Conditions where the myelin or the structure of the axon are disrupted, such as trauma, tumors, and inflammation reduce anisotropy, as the barriers are affected by destruction or disorganization.
Anisotropy is measured in several ways. One way is by a ratio called "fractional anisotropy" (FA). An anisotropy of "0" corresponds to a perfect sphere, whereas 1 is an ideal linear diffusion. Well-defined tracts have FA larger than 0.20. Few regions have FA larger than 0.90. The number gives information of how aspherical the diffusion is but says nothing of the direction.
Each anisotropy is linked to an orientation of the predominant axis (predominant direction of the diffusion). Post-processing programs are able to extract this directional information.
This additional information is difficult to represent on 2D grey-scaled images. To overcome this problem a color code is introduced . Basic colors can tell the observer how the fibers are oriented in a 3D-coordinate system: This is termed an "anisotropic map". The software could encode the colors in this way:
- Red indicates directions in the X axis: right to left or left to right.
- Green indicates directions in the Y axis: posterior to anterior or from anterior to posterior.
- Blue indicates directions in the Z axis: foot-to-head direction or vice versa
Notice that the technique is unable to discriminate the "positive" or "negative" direction in the same axis.
References
- ^ Filler, Aaron (2010). "The History, Development and Impact of Computed Imaging in Neurological Diagnosis and Neurosurgery: CT, MRI, and DTI". Internet Journal of Neurosurgery. 7 (1).
{{cite journal}}: External link in|journal=
External links
- MedINRIA: A freeware (non-commercial use only) for DTI tractography
- 3D Slicer: Free software with DTI tractography calculation and visualization (BSD License)
- TrackVis: DTI tractography visualization software
- Dipy: Diffusion Imaging in Python
