User:Kinkreet/Protein Science/Photosynthesis
Introduction
[edit]Respiration
- C6H12O6 (aq) + 6 O2 (g) → 6 CO2 (g) + 6 H2O (l) + heat
- oxygen + glucose → carbon dioxide + water + energy
- ΔG = -2880 kJ per mole of C6H12O6
Photosynthesis
- 2n CO2 + 2n H2O + photons → 2(CH2O)n + 2n O2
- carbon dioxide + water + light energy → carbohydrate + oxygen
- ΔG = +2870 kJ per mole of C6H12O6
Every year, >1018 kJ of free energy are stored using photosynthesis, which corresponds to storing ~1011 tons of carbon into carbohydrates.
Rhodopsin-based photosynthesis
[edit]Photons provide energy for rhodopsin to transport protons across a membrane. The built up proton gradient provides energy to drive ATP synthase to produce ATP from ADP and Pi.
Evolution of Photosynthesis
[edit]The first anoxygenic photosynthetic prokaryotic bacteria emerged ~3.2 billion years ago, these bacteria use energy from photons to fix carbon but did not produce any oxygen. The first oxygenic photosynthetic bacteria emerged ~2 billion years ago, and subsequently created the oxidising atmosphere.
All reaction centres derived from a common ancester.[1]
History
[edit]It was previously assumed that the energy derived from photons was used to reduce CO2 to carbon and oxygen, where the carbon is combined with water to give carbohydrates, and oxygen is released into the atmosphere.
However, this presumption was overturned in 193?, when van Niel showed that Purple Sulfur Bacteria uses H2S in photosynthesis to generate sulphur
2H2S + CO2 → (CH2O) + H2O + 2S
Because H2S are similar chemically, van Niel proposed that the oxygen released in oxygenic photosynthesis comes from the water and not the carbon dioxide. The energy from the photon is used to oxidize H2A to A and 2[H] in the light reaction, where the generated reducing agent is used to reduce CO2 to carbohydrates during the dark reaction. It is now known that the light reactions produce reducing power (in the form of NADPH, which carries the protons), ATP and O2. O2 is released; ATP and NADPH provides the energy and reducing power, respectively, to fix CO2 into carbohydrates.
The general equation of photosynthesis can be written as: 2H2A + CO2 → (CH2O) + H2O + 2A, where A is any element.
Chloroplast
[edit]
1. outer membrane
2. intermembrane space
3. inner membrane (1+2+3: envelope)
4. stroma (aqueous fluid)
5. thylakoid lumen (inside of thylakoid)
6. thylakoid membrane
7. granum (stack of thylakoids)
8. thylakoid (lamella)
9. starch
10. ribosome
11. plastidial DNA
12. plastoglobule (drop of lipids)
Chloroplasts are organelles found in higher plants, and is the centre for photosynthesis. They are thought to have derived from cyanobacteria forming an endosymbiotic relationship with eukaryotes. Chloroplast takes a ellipsoidal structure ~5μm in length. It is made up of a phospholipid outer membrane (derived from plants) surrounding an inner membrane (remnants from prokaryotes/bacteria); inside the membrane lies the stroma, which is analogous to the cytosol of bacteria, and contains small circular DNA and ribosomes, although most of the proteins are synthesized by the host and transported into the chloroplast. In the stroma lies an extensive layered membrane structure called thylakoids, which stacks up to form grana. The thylakoid membrane is home to the redox centres, and creates a barrier to which proton gradient is stored inside the thylakoid space or lumen. The grana are connected by stromal lamella. Most of the photosystem II tends to be localized in the granal regions, whereas PSI and ATP synthase tend to localize in the stromal lamella; cyt b6f is even distributed across the thylakoid membrane. In plants, the ratio of PSI:PSII are around 10:1.
Recent studies have shown that chloroplasts can be interconnected by tubular bridges calledstromules, formed as extensions of their outer membranes.[2][3] Chloroplasts appear to be able to exchange proteins via stromules,[4] and thus function as a network.
Oxygenic photosynthesis
[edit]Electron transport begins with the splitting of water molecules using Photosystem II to generate electrons, which get passed sequentially through plastoquinone (PQ), cytochrome b6f (Cyt b6f), plastocyanin (PC), Photosystem I (PSI) and PSI-bound ferredoxin (Fd). Finally, it is used by ferredoxin–NADP+ oxido-reductase in the stromal matrix to produce NADPH, which is required as part of the Calvin cycle.
There are two types of oxygenic photosynthesis - linear and cyclic. They exists to control the balance between the production of ATP (source of energy) or NADPH (reducing power). Cyclic photophosphorylation is more prominent when the ratio of NADPH:NADP+ is high, and thus the reducing power is high already, so the energy derived is better stored as ATP. In both types of photosynthesis, water is the source of electrons.
Photosystem II
[edit]| Overall reaction | |
|---|---|
| Lumen | 2H2O → O2 + 4 H+ |
| Stroma + Membrane | 4 hv + 4 H+ (in stroma) + 2 Plastoquinone (PQ, in membrane) → 2 Plastoquinol (PQH2, in membrane) |
| Net movement of 4 proton from the stroma to the thylakoid lumen, and reduction of plastoquinone to plastoquinol | |
Photosystem II is a complex consisting of 16 integral membrane proteins, 3 soluble proteins, 36 chlorophylls, 7 carotenoids, 2 pheophytins, 2 haems, 2 plastoquinones and 1 manganese cluster in an oxygen-evolving water-spliting centre.
Light harvesting
[edit]| Phycobilisome protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 The layout of protein subunits in a phycobilisome. | |||||||||
| Identifiers | |||||||||
| Symbol | Phycobilisome | ||||||||
| Pfam | PF00502 | ||||||||
| InterPro | IPR001659 | ||||||||
| SCOP2 | 1cpc / SCOPe / SUPFAM | ||||||||
| |||||||||
Light-harvesting Chl b-binding (LHC II) proteins associated with Photosystem II first absorb photons, where the energy derived is used to split water into molecular oxygen, protons and electrons. The protons enter the lumen of the thylakoid which builds up a concentration gradient.
In higher plants and green algae, the LHCs are arranged within the thylakoid membrane, in cyanobacteria, the LHCs are soluble proteins arranged in the stromal matrix as long rods. Light harvesting pigments include chlorophyll a, chlorophyll b, β-carotene, other chlorophylls and carotenoids.
In cyanobacteria, red algae and glaucophytes, allophycocyanin, phycocyanin, phycoerythrin and phycoerythrocyanin (collectively known as phycobiliproteins) form a phycobilisome, which is used as the antenna used in PSII. Phycocyanin is simply chlorophyll set out in a line, and phycoerythrin have a saturated C-C bond at one position, and this changes the absorption spectra dramatically.
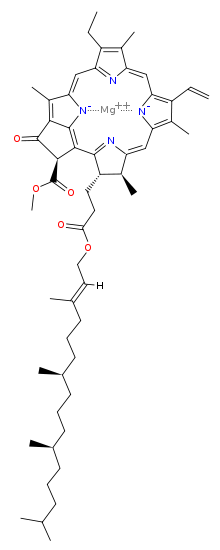
The most prominent pigment is chlorophyll a, followed by b. Chlorophylls a, b and d are identical apart from single aldehyde groups replacing the alkyl group. By changing the substituent group, the absorption spectra are changed.
Chlorophylls a, b and d are tetrapyrroles which are synthesized using a similar pathway as the Protoporphyrin IX (Fe+) group in haemo- and myoglobin. N.B. The pyrroles co-ordinate a central magnesium ion and not manganese, which is used to split water.
PSII requires excitation every milisecond, having only chlorophylls will not be enough to produce such high output. Therefore, the light harvesting mechanism contains a reaction center chlorophyll surrounded by antenna pigment molecules, which are excitationally-coupled to each other, and so the excited electrons can resonate from one antenna pigment to another, until it reaches the reaction centre. This way, energy from many photons are funneled down to one place, and the collective energy is enough to drive the splitting of water.
Energy can be transferred from one antenna pigment to another using resonance transfer, or electron transfer. Resonance transfer is where an electron is excited, by a photon, to its excited state. As it de-excites, it releases another photon which is absorbed by a neighbouring electron on another molecule; this recurs and energy is transferred from one molecule to another. In electron transfer, a.k.a. photoinduced charge separation, an electron is excited to an excited state (i.e. an orbital of higher energy). But the neighboring molecule has an orbital of equal or lower energy which overlaps or is in close proximity with our excited electron, at which point the excited electron will move from its original molecule to this neighbouring molecule. If the neighboring molecule have no vacancy at the lower energy orbitals, then the electron will remain at the excited state. The excitation energy is mostly converted into charge separation.
Note: When an electron is excited, it can be donated to another molecule, and thus acts as a good reducing agent; however, it also leaves a vacant space at the orbital it got excited from, to which it can accept electrons, and thus can also act as a good oxidizing agent.
Water splitting using the Oxygen-evolving complex
[edit]The oxygen-evolving complex (OEC) store up the energy required to oxidize water in the form of successive positive charges. There are 5 stages as described by Kok[5]: S0 to S4+. Kok used flashes of light to investigate the number of photons required to provide energy for the oxidation to occur. He found that there is a large peak in oxygen yield every fourth flash, this suggests that the OEC stores energy from four photons for each cycle of water oxidation. This fits in well because there are 4 individual reaction centres (Mn), or that one manganese can have 4 oxidation states (II - V).
When a photon is absorbed, energy is transferred to P680, which donates it electrons to an acceptor. P680 is now electronegative (-1.2V) and removes electrons from the OEC. The OEC moves from the S0 state to the S1 state, both of which are stable in the dark. Subsequent photon absorptions leads to the 'promotion' of the OEC to a higher state, up to the S4+ state. The S4+ state is unstable and will oxidize water and return the OEC to the ground state. The flashes must occur in the microsecond scale, and must follow closely from each other, as the relaxation times from one state to a lower one is ~200-400μs. This cycle is also known as the Kok cycle, or S cycle (S for storage). Each cycle releases 4 electrons and 4 protons, as well as generate one molecule of molecular oxygen.
The structure of the OEC has 4 manganese to one calcium, possible in the configuration Mn4Ca1OxCl1–2(HCO3)y.
Electron transfer
[edit]Electrons are transferred from Mn4Ca complex (OEC) to TyrZ+ (YZ+) to P680+ (named because it absorbs at 680nm). P680 is the most power oxidizing agent known, and it will take up all the electrons given out by the OEC. Electrons are then passed to pheophytin (Phe), QA, via Fe to QB and finally to PQH2.
Many electron carriers are required because electrons cannot be transferred across large distances. It has shown a general trend that the rate of transfer decays proportional to the exponent of the distance. Given that the membrane is 5-10μM thick, to transfer electrons using just one donor and one acceptor would take a long time
Cytochrome b6f
[edit]
Cytochrome b6f is light-independent protein with close structural and functional homologue of respiratory complex III. In its homodimeric form, it consists of 4 large subunits: cyt f (33kDa, contains c-type haem), cyt b6 (23kDa, contains 2 b-type haems), Rieske-type Fe-S protein (20kDa, contains one [2Fe-2S] cluster, and subunit IV (17kDa)
In the thylakoid membrane, the PQH2 is used to reduce quinone to QH2 (a.k.a. coenzyme Q : cytochrome c — oxidoreductase, cytochrome bc1complex, complex III), which enters cytochrome b6f. PQH2 is oxidized back to PQ. QH2 is oxidized to generate two protons which is released into the lumen; two more protons are pumped in from the stroma. QH2 oxidation also generate electrons which is passed through the Rieske-type Fe-S protein, Fe of cyt f and finally to plastacyanin to reduce it.
Plastacyanin (99AA, ~10.5kDa) is a monomeric, copper-containing protein involved in electron-transfer. It transports electrons from cyt f to P700+ from PSI.
Photosystem I
[edit]PSI is a multisubunit complex, consisting of more than 13 subunits. It is a trimer in cyanobacteria, and a monomer in eukaryotes.
By the time electrons reach PSI, much of the energy have already been lost, and so PSI is required to be 'boost' the energy of the electrons, by absorbing photons.
There are 11 reaction centres (6 chlorophylls, 2 phylloquinones, 3 [4Fe-4S] clusters) in PSI and each is bound by a transmembrane helix. One of the reaction centres is a special pair of chlorophyll a molecules called P700, and it acts as the primary electron donor. Energy derived from reaction centres absorbing photons are transferred onto P700, which donates its electron to A0; this makes P700 electronegative enough (-1.3V) to remove an electron from plastocyanin (Cu+). A0 acts as the primary electron acceptor. (The oxidized plastocyanin (Cu2+) can now return to cyt b6f for the next round of electron transport)
Electrons are then passed from A0 to A1 and finally to a set of [4Fe-4S] clusters on ferredoxin, the final electron acceptor of the PSI system.
Linear photosynthesis
[edit]Ferredoxin is soluble and diffusible in the stroma. In linear (or non-cyclic) photosynthesis, ferredoxin reduces NADP+ to generate NADPH, which is used as a source of reducing power in the carbon fixation reactions.
Two photons, two reduced plastocyanine are required for reduction of 2 ferredoxin molecules. The two reduced ferredoxins can then react with one NADP+ and one H+ to generate NADPH + H+ and 2 oxidized ferredoxin; this reaction is carried out by ferredoxin:NADP+ reductase (FNR). FNR requires a flavin cofactor, FAD, which is the entity which accepts the electron, one at a time from two molecules of Fdred, going through the semiquinone to the fully reduced form.
- 2 reduced ferredoxin + NADP+ + 2H+ 2 oxidized ferredoxin + NADPH + H+
Fdred is a strong reductant, but it can only donate one electron. NADPH can donate 2 electrons, and so is more widely used.
PSI is homologous to PSII and other reaction centres where it takes in light energy to produce reducing power. Whereas PSII generates plastoquinone, PSI generates reduced ferredoxin; PSI also does not oxidize water.
In linear photosynthesis, energy from 8 photons are used to generate one molecular oxygen, 2 NADPH and 3 ATP.
Cyclic photosynthesis
[edit]In cyclic photosynthesis, ferredoxin transfers electrons back to cyt b6f, which will use the electrons to pump protons across the membrane (using the Q cycle). This increases the proton gradient which increases rate of ATP synthesis.
In cyclic photosynthesis, energy from 2 photons are used to generate 2 ATP.
ATP synthase
[edit]CF0F1 ATP synthase is similar to mitochondria ATP synthase, and uses the dissipation of 3 protons from the lumen to the stroma, to synthesize one ATP from ADP + Pi. The proton gradient is generated by taking protons from the stroma to the lumen (4 from PSII and 4 from cyt b6f), and maintained because the thylakoid membrane is impermeable to protons. The pH of the lumen is typically ~pH4, whereas in the stroma it's ~pH8, so the difference can be up to 3.5 units.
Regulation
[edit]The apparatus used to convert light energy to chemical energy are themselves susceptible to photodamage. Nonphotochemical quenching (NPQ) is a mechanism used by plants and eukaryotic algae, by which the excitation energy is dissipated as heat, preventing damage by light energy.
Notes
[edit]- Light-harvesting Chl a-binding (LHC I)
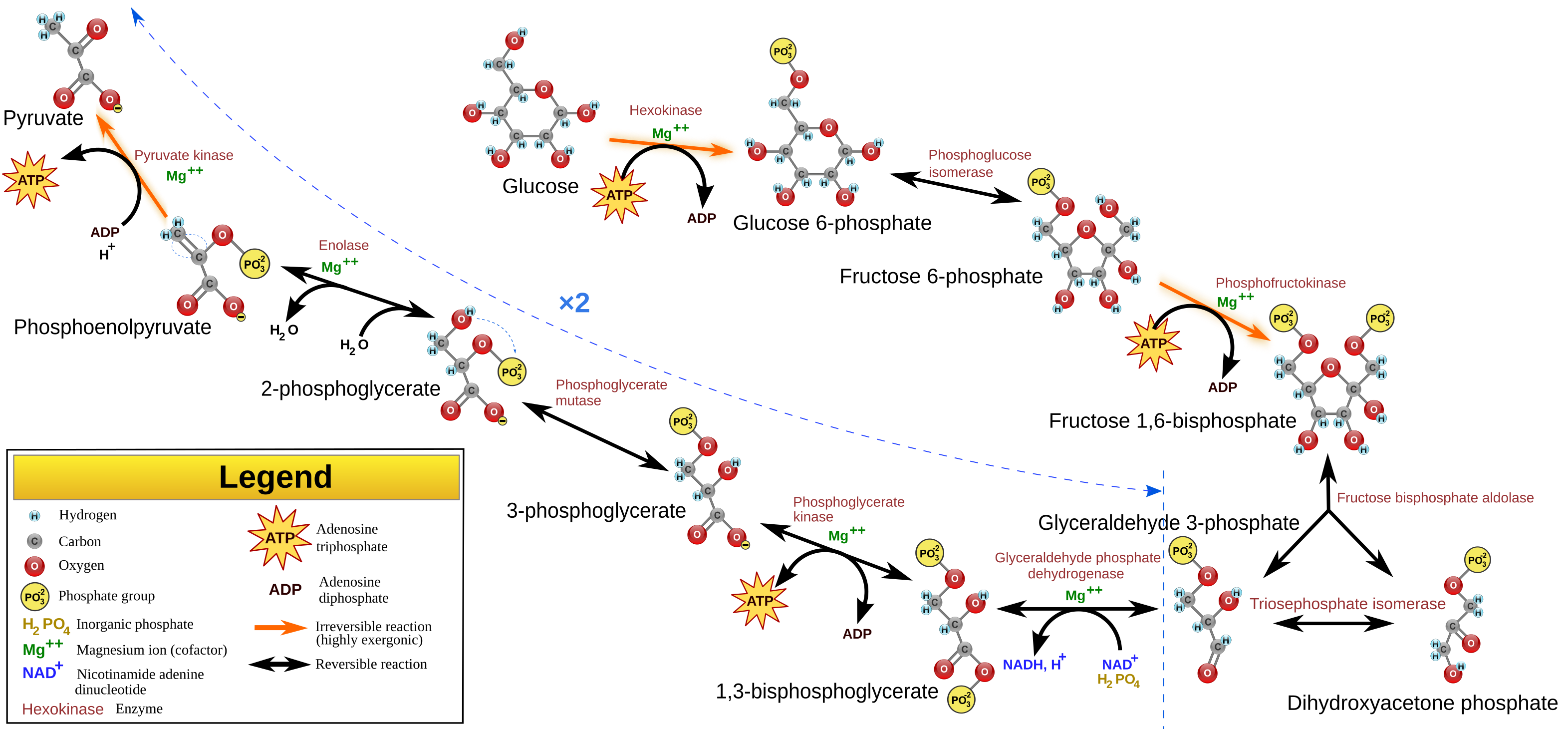
Calvin Cycle
[edit]The Calvin cycle takes in carbon dioxide from the atmosphere, and using the reducing power of NADPH and energy in the form of ATP, created using the light reaction, to drive the reduction of carbon dioxide to glyceraldehyde-3-phosphate (G3P).
Ribulose-1,5-bisphosphate carboxylase-oxygenase (Rubisco) incorporates CO2 into Ribulose-1,5-bisphosphate to give two molecules of 3-phosphoglycerate The NADP+ and ADP generated from this cycle is fed back to the chloroplast where they can be regenerated to form NADPH and ATP.
Fixation
[edit]Reduction
[edit]Regeneration
[edit]Notes
[edit]- Photosynthetic efficiency
- Co2 and O2 cycle in the cell
- RuBisCo
- Dependence on environmental conditions and regulation
- Photorespiration
- C4 and CAM
- Carbon concentrating mechanisms
- Other autotrophic carbon-fixing cycles - 3HP cycle
Climate Change
[edit]An equilibrium of CO2 is achieved by photosynthetic organisms reducing CO2 into carbohydrates, countered by microbes, plants and animals metabolizing the carbohydrates, and producing CO2. The carbohydrates can also be stored as fossil fuels, which when combusted, releases its carbon into the atmosphere as CO2.
Climate change can be measured against the concentration of CO2 in the atmosphere. The longest continuous record of atmospheric CO2 levels is done at the Mauna Loa Observatory in Hawaii, which is as accurate as an indication can be, because Hawaii is surround by the sea, which acts as a buffer as it can dissolve CO2 into carbonic acid (H2CO3), and so the changes observed (a continuous increase) is a global phenomenon, and not caused by local effects. From these recordings, it can be shown that CO2 levels increase by 25% since 1958 (when the first recording was made). As of May 2012, CO2 levels stand at 396.78 ppm, a 2.6 ppm increase from May 2011.
Because CO2 is a greenhouse gas, an increased level of CO2 means an increase in global temperature, causing greater differences in climate, increasing days of droughts and floods. However, it may also fertilize plants, as plants take in CO2 as a source of biomass. According to ESPACE (European Spatial Planning: Adapting to Climate Events), doubling the levels of CO2 for wheat (single genotype) increased the rate of photosynthesis by 25-58%, and an increase in crop yield of 35%.
Energy efficiency
[edit]Most of the energy from the photons that befalls the photosynthetic organisms is loss during the reactions of photosynthesis. Mitochondrial respiration using 40% of the energy derived, the photosystems only absorb between 400-600 nm, and so photons of other wavelengths are no utilized, and so 58% of the energies are loss; Calvin cycle wastes 66% of its input energy, and photorespiration results in a 30% loss. Overall, the efficiency of plants is less than 1%, with the most significant loss of energy comes from the Calvin cycle.
Calvin Cycle
[edit]The Calvin cycle is the light-independent reaction of photosynthesis, and uses the reducing power and energy produced in the light-dependent reactions to fix carbon. It usually occurs at night, in the stromal matrix of chloroplast. Although it does not require energy from photons, it is accelerated in the presence of light because more reducing power and energy is available from the light-dependent reactions to drive carbon fixation.
Non-photosynthetic organism, such as some archaea, uses the Calvin cycle to fix carbon, but instead of using energy from photons to generate energy, it have a chemical means of energy generation. The fixation of carbon involves adding CO2 to a larger molecule, which is retained in the body as biomass.
There are relatively few carbon fixation pathways compared to other pathways, because the carbon fixation pathway is complicated and difficult to evolve to.
History
[edit]The original Calvin cycle was elucidated between 1946 and 1953 by Melvin Calvin, Andrew Benson and James Bassham.
It followed from Ruben and Kamen's experiment in 1941. They grew algae in a lollipop-shaped flask, providing it with light and air. They then injected H14CO3 into the suspension. Periodically after injection, samples were taken and added to hot methanol, which stopped the reaction. As 14C is radioactive, these extracts can be ran on 2D paper chromatography, with two different solvents (phenol:H2 and butanol:propionic acid), to identify by autoradiography against controls, the compounds present with 14C. They noted the order at which the different compounds appear, and assumed that the 14C was passed down from one to the other, thus allowing them to draw a pathway outlining the exchange of the 14C.
Overview
[edit]The Calvin cycle can be split into three phases: carbon fixation, reduction, and regeneration.
Carbon fixation is where RuBisCo (Ribulose bisphosphate carboxylase) attaches carbon from CO2 to ribulose bisphosphate (RuBP, a 5-carbon sugar). The product of this reaction is ????, but it is unstable and immediately splits to form two molecules of 3-phosphoglycerate (3PG).
The 3-phosphoglycerate is converted to glyceraldehyde-3-phosphate (G3P) using ATP and NADPH2 derived from the light-dependent reactions. Phosphoglycerate kinase uses 1 ATP to phosphorylate 3PG to 1,3-bisphosphoglycerate, after which GAPDH uses NADPH2 to reduce that to G3P. G3P is a pivotal intermediate of the glycolysis pathway, and thus can be used to synthesize other sugars. Therefore, G3P is the output of the calvin cycle.
5 out of 6 G3P generated is used to regenerate RuBP, only one is exported to synthesize sugars. In regeneration, 3 ATP and 5 G3P is used to regenerate 3 RuBP. RuBP must be regenerated to accept another CO2 molecule.
As CO2 is a one-carbon molecule, and G3P is a three-carbon molecule, three molecules of CO2 must be fixed to produce one molecule of G3P. For every G3P synthesized, 9 ATPs and 6 NADPH2 is utilized.
The carbon fixation and reduction step occurs in a linear manner; regeneration, however, is much more complex. This is because 3-carbon sugars must be used to regenerate a 5-carbon sugar, and so there are many interlinking steps involved.
Fixation
[edit]RuBisCo is made up of 8 large subunits (55kDa each, arranged into 4 dimers) and 8 small subunits (13kDa), making up a total size of ~540kDa. The small subunits are not universally essential and act only to enhance the catalytic activity of the large subunits; all the active and regulatory sites lie on the large subunits. It require magnesium ions (Mg2+) at the active site, and is activated upon binding to one of a non-substrate CO2.
It is located close to the stromal surface of the thylakoid membrane. It catalyzes the first reaction in the Calvin cycle, carboxylating ribulose bisphosphate (RuBP, a 5-carbon sugar) using CO2. The product of this reaction is a labile 6-carbon compound, but it is unstable and immediately splits to form two molecules of 3-phosphoglycerate (3PG).
RuBisCo is both non-specific and very slow, able to bind to and catalyse oxygen as well as carbon dioxide, meaning it can both carboxylate and oxygenate RuBP. It catalyzes ~3 reactions a second (a 'typical' enzyme can catalyze ~1000 reactions per second), which is really slow. Because its poor specificity and slow turnover rate, it is required in high quantity. Indeed, it is the most abundant enzyme in nature (about half the soluble proteins in a leaf is RuBisCo). Its reaction rate is high with higher pHs, and is also dependent on products from the light reactions (ATP and NADPH2), the stromal concentration of Mg2+ etc. RuBisCo activase is a 43kDa protein which is activated by ATP. It activates RuBisCo. RuBisCo can also misfire and produce xylulose 1,5-bisphosphate (XuBP) and 3-keto-arabinitol 1,5-bisphosphate (KABP) which binds tightly to RuBisCo activase and inactivates it. Binding of these inhibitors leads to a loop on RuBisCo activase to be extended and covers the active site. Some plants make other inhibitors on purpose, such as 2-carboxy-D-arabinitol-1-phosphate (CA1P), which binds to RuBisCo; it is removed by RuBisCo activase and also degraded by CA1P phosphatase.
Mechanism
[edit]First, RuBP carboxylase activase is used to add a non-substrate CO2 to lysine-201 to form a carbamate, allowing it to take part in coordinating a magnesium ion along with an aspartate and a glutamate residue. Once RuBisCo is activated, RuBP enters into the active site. The phosphate groups are stabilized by hydrogen bonds by positively-charged lysine, argnine and histidine residues. The positively-charged magnesium ion stabilizes the double-bonded oxygen and hydroxide at the 2 and 3 positions, respectively.
Because magnesium is positively-charged, some of the electrons in the double bond moves towards the magnesium; this forms a double bond between C2 and C3, resulting in the deprotonation at C3 to give an enediolate intermediate. CO2 now enters. The double bond of RuBP nucleophilic attack the carbon, while the Mg<sup2+ ion stabilizes the oxygen on CO2 by hydrogen bonding. After the nucleophilic attack, water rapidly attacks the intermediate to give a 6-carbon compound, which quickly dissociates into 3PG.
Reduction
[edit]The 3-phosphoglycerate is converted to glyceraldehyde-3-phosphate (G3P) using ATP and NADPH2 derived from the light-dependent reactions. Phosphoglycerate kinase uses 1 ATP to phosphorylate 3PG to 1,3-bisphosphoglycerate, after which GAPDH uses NADPH2 to reduce that to G3P. The second step runs parallel to the gluconeogenesis step, however, in gluconeogenesis, glyceraldehyde phosphate dehydrogenase uses NADH/NAD+rather than NADPH. The resulting GAP can be used to carry on the rest of gluconeogenesis to give a range of hexoses. These hexoses can be used to make carbohydrates, which can be stored. Starch (or amylose) is a polymer of glucose. It is similar to glycogen but have less α-1,6-glycoside linkages, and thus is less branched. It is used mainly by plants and stored in chloroplasts. Sucrose is a dissacharide of glucose and fructose, and is synthesized in the cytosol. In plants, because hexose phosphate cannot be transported across the chloroplast membrane, it is exported as a triose phosphate (G3P), and the incorporation into hexoses to make sucrose is carried out in the cytosol.
Sugar synthesis must be controlled as to ensure the levels of G3P do not get depleted or else regeneration of RuBP will be slowed down until glycolysis reproduce these triose sugars back.
Regulation
[edit]The Calvin cycle activity changes depending on the environmental condition the organism is in. In photosynthetic organisms, the presence of light determines how much ATP and NADPH is present to drive carbon fixation; stromal levels of Mg2+ and pH determines the activity of RuBisCo. RuBisCo favours a higher pH, and the plant adapts to this; when light is shown on chloroplast, Mg2+ comes out from the thylakoid lumen into the stroma and activates RuBisCo, while protons are pumped by the light reactions into the thylakoid lumen, reducing pH in the stroma.
Thioredoxin is a 12kDa protein which contains a redox active cystine/two cysteine. When it is reduced activates many enzymes. Reduced ferrodoxin from the light reaction reduces thioredoxin via ferrodoxin-thioredoxin reductase, which can then move on to activate (by reducing) Calvin cycle enzymes.
CP12 is a small disordered protein with a pair of conserved cysteines. It binds to phosphoribulokinase and GAPDH, inhibiting the regeneration of RuBP as well as the generation of G3P, choking both sides of carbon fixation. When NADPH from the light reaction is present, it releases CP12 leading to activation.
Photorespiration
[edit]Because RuBisCo have a low specificity, and thus cannot distinguish between CO2 and o2, and thus will oxygenate RuBP as well as carboxylate it. Many other carboxylases use hygrogencarbonate (HCO3) instead to increase specificity, however, because RuBisCo is such an important enzyme, it is conserved and thus have not evolved to the specificity of other carboxylases.
Instead of the double bond of the enediolate intermediate attacking the carbon on CO2, O2 is added onto the O- group on C2. Upon hydrolysis by water, instead of yielding two molecules of G3P, one G3P and one phosphoglycolate is produced. Phosphoglycolate is a two-carbon compound and is toxic. This results in a net loss of 2 carbons and the production of a toxic molecule. A salvage mechanism is used to remove the toxicity, converting two molecules of phosphoglycolate to 1 G3P and releasing a molecule of CO2.
Photorespiration occurs ~4x slower than carbon fixation, and so 20% of all reactions catalyzed by RuBisCo is photorespiration. This effect is more prominent at higher temperatures. Photorespiration is a huge cost as it wastes CO2, ATP and NADPH, it has no confirmed metabolic function. Some suggests that it is an imperfection in evolution, while some suggests that it protects the photosynthetic apparatus from photooxidative damage when CO2 levels are down and the energy cannot be dissipated; using up the oxygen removes it from the stroma to prevent the formation of oxygenic free radicals.
C3 and C4
[edit]The C4 Calvin cycle is where carbon dioxide is concentrated near the Calvin cycle enzymes to minimize the rate of photorespiration. As oxygenase activity increases more than the carboxylase activity with higher temperatures, it becomes a bigger problem. Therefore, C4 plants are mainly found in tropical plants and hot environments.
The anatomy in C4 plants are different to those in C3 plants. In C3 plants, carbon dioxide is able to enter through the stomata into a spongy mesophyll layer and into the vein, where the bulk of the Calvin cycle enzymes resides. In C4 plants, the carbon dioxide is concentrated onto bundle sheath cells surrounding the vein, and thus increases the level of CO2 in these areas, pushing for carboxylation rather than oxygenation. CO2 is concentrated as mesophyll cells (in contact with the air and carries out much of the light reactions) transfer four-carbon compounds such as malates and oxaloacetates to the bundle sheath cells. Phosphoenolpyruvate (PEP) is condensed with HCO3 (derived from CO2 using carbonic anhydrase) to give oxaloacetate, which is reduced by NADPH to malate. This malate is then exported into the chloroplast of the bundle sheath cells, where it is decarboxylated using NADP+ into pyruvate and CO2. CO2 enters the Calvin cycle to be fixed, while the pyruvate is exported back into the mesophyll cells, where ATP hydrolysis to AMP is used to phosphorylate and regenerate phosphoenolpyruvate. The AMP requires two ATP hydrolysis to regenerate one ATP. Thus, the concentration of CO2 to the bundle sheath cells uses 2 ATP per CO2 moved. This is expensive but the concentration of CO2 in the bundle sheath cell is 20x that in the mesophyll, minimizing the effects of photorespiration.
C3 6 CO2 + 18 ATP + 12 NADPH + 12 H2O → C6H12O6 + 18 ADP + 18 Pi + 12 NADP+ + 6 H+
C4 6 CO2 + 30 ATP + 12 NADPH + 12 H2O → C6H12O6 + 30 ADP + 30 Pi + 12 NADP+ + 18 H+
Although more energy is used to fix one carbon, because C4 plants are more efficient in keeping CO2, its stomata do not need to be opened for as long as C3 plants. This way, they lose less water and become more productive. This means C4 plants are some of the most productive plants known; examples includes sugar cane and maize. Efforts have been put into making wheat and rice plants into more of a C4 plant, to improve productivity.
C4 plants are observed a different points in the phylogenetic tree, and thus it has evolved multiple times[6]
Crassulacean acid metabolism
[edit]Crassulacean acid metabolism (CAM) is another carbon fixation pathway evolved in some plants as an adaptation to arid conditions. Examples includes cacti, pineapple and plants of the Crassulaceae family. CAM plants tend to be slow growing.
These plants open their stomata at night to allow CO2 to enter. Temperature is low at this point and so it minimizes the loss of water. CO2 is stored as malic acid in the vacuole of the cell overnight. During the day time, the stomata is closed and water cannot escape; malic acid moves out from the vacuole to be decarboxylated into pyruvate and CO2, which enters into the Calvin cycle to be fixed.
Cyanobacteria
[edit]C4 and CAM are adaptations in the carbon fixation pathways seen in plants to concentrate CO2 to give a higher rate of carbon fixation. In cyanobacteria, similar mechanisms exists.
Energy is used to pump CO2 and HCO3 into the cyanobacteria. pH is regulated using pumps to ensure all CO2 is converted into the bicarbonate anion (HCO3-) inside the cytoplasm-equivalent of the cyanobacteria. HCO3- is then transported into the carboxysome. The carboxysome is a bacterial viral-like proteinous compartment used to concentrate CO2 to enhance fixation. It has a high level of RuBisCo and carbonic anhydrase for this purpoes (the HCO3- is converted into CO2 using carbonic anhydrase).
The structure of the carboxysome is a regular icosahedron (30 edges and 12 vertices) shell, with faces made up of hexameric subunits, and the vertices made up of pentameric subunits[7]. Small pores are present through the hexamers and may serve as the route for diffusion of substrates (bicarbonate and ribulose-1,5-bisphosphate, but not oxygen) and products (3-phosphoglycerate) into and out of the carboxysome.
Alternative Carbon Fixation Pathways
[edit]Although not many exists (as it is hard to evolve), alternative carbon fixation pathways have been found, and includes: reductive citric acid cycle, Wood-Ljungdahl pathway (used by anaerobic chemoautotrophs)[8], Archaeal cycles[9] and the 3-hydroxypropionate cycle. Unlike the Wood-Ljungdahl pathway which is extremely oxygen-sensitive, the 3-hydroxypropionate cycle is not. It is found in the bacteria Chloroflexus aurantiacus, a photosynthetic green non-sulfur bacteria found in hot springs. It does not use water as the electron donor, but others including H2S, and thus gives out no oxygen. The carbon is fixed in anaerobic conditions via the 3-hydroxypropionate cycle (rather than the Calvin cycle). This cycle is more productive than the C4 pathways.
| Carbon fixation pathway | ATP | NADPH |
|---|---|---|
| C3 Calvin cycle | 9 | 6 |
| C4 Calvin cycle | 15 | 6 |
| 3-hydroxypropionate cycle | 11 | 6 |
References
[edit]- ^ Hohmann-Marriott, Martin F.; Blankenship, Robert E. (2011). "Evolution of Photosynthesis". Annual Review of Plant Biology. 62: 515–48. doi:10.1146/annurev-arplant-042110-103811. PMID 21438681.
- ^ Köhler RH, Hanson MR (1 January 2000). "Plastid tubules of higher plants are tissue-specific and developmentally regulated". J. Cell. Sci. 113 (Pt 1): 81–9. doi:10.1242/jcs.113.1.81. PMID 10591627. Archived from the original on 2010-11-18.
- ^ Gray JC, Sullivan JA, Hibberd JM, Hansen MR (2001). "Stromules: mobile protrusions and interconnections between plastids". Plant Biology. 3 (3): 223–33. doi:10.1055/s-2001-15204.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR (June 1997). "Exchange of protein molecules through connections between higher plant plastids". Science. 276 (5321): 2039–42. doi:10.1126/science.276.5321.2039. PMID 9197266. Archived from the original on 2010-11-18.
{{cite journal}}: CS1 maint: date and year (link) CS1 maint: multiple names: authors list (link) - ^ Kok, Bessel; Forbush, Bliss; McGloin, Marion (1970). "Cooperation of Charges in Photosynthetic O2Evolution–I. A Linear Four Step Mechanism". Photochemistry and Photobiology. 11 (6): 457–75. doi:10.1111/j.1751-1097.1970.tb06017.x. PMID 5456273. S2CID 31914925.
- ^ Sage, R. F.; Christin, P.-A.; Edwards, E. J. (2011). "The C4 plant lineages of planet Earth". Journal of Experimental Botany. 62 (9): 3155–69. doi:10.1093/jxb/err048. PMID 21414957.
- ^ Kerfeld, C. A.; Sawaya, MR; Tanaka, S; Nguyen, CV; Phillips, M; Beeby, M; Yeates, TO (2005). "Protein Structures Forming the Shell of Primitive Bacterial Organelles". Science. 309 (5736): 936–8. doi:10.1126/science.1113397. PMID 16081736. S2CID 24561197.
- ^ Ragsdale, SW; Pierce, E (2008). "Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1784 (12): 1873–98. doi:10.1016/j.bbapap.2008.08.012. PMC 2646786. PMID 18801467.
- ^ Berg, IA; Kockelkorn, D; Ramos-Vera, WH; Say, RF; Zarzycki, J; Hügler, M; Alber, BE; Fuchs, G (2010). "Autotrophic carbon fixation in archaea". Nature Reviews. Microbiology. 8 (6): 447–60. doi:10.1038/nrmicro2365. PMID 20453874. S2CID 16059500.