Cancer biomarker: Difference between revisions
→Recurrence: - Added references to the recurrence section and more details on the OncotypeDx |
|||
| Line 35: | Line 35: | ||
====Recurrence==== |
====Recurrence==== |
||
Lastly, cancer biomarkers offer value in predicting or monitoring cancer [[relapse|recurrence]]. The [http://www.oncotypedx.com/ Oncotype DX®] breast cancer assay is one such test used to predict the liklihood of breast cancer recurrence. Thist test is intended for women with [[Cancer staging|early-stage]] (Stage I or II), node-negative, [[estrogen receptor]]-positive (ER+) invasive breast cancer who will be treated with [[hormone therapy]]. Oncotype DX looks at a panel of 21 genes in cells taken during tumor [[biopsy]]. The results of the test are given in the form of a recurrence score that indicates liklihood of recurrence at 10 years.<ref>{{cite journal|last=Lamond|first=NW|coauthors=Skedgel, C; Younis, T|title=Is the 21-gene recurrence score a cost-effective assay in endocrine-sensitive node-negative breast cancer?|journal=Expert review of pharmacoeconomics & outcomes research|date=2013 Apr|volume=13|issue=2|pages=243-50|pmid=23570435}}</ref> <ref>{{cite journal|last=Biroschak|first=JR|coauthors=Schwartz, GF; Palazzo, JP; Toll, AD; Brill, KL; Jaslow, RJ; Lee, SY|title=Impact of Oncotype DX on Treatment Decisions in ER-Positive, Node-Negative Breast Cancer with Histologic Correlation.|journal=The breast journal|date=2013 May|volume=19|issue=3|pages=269-75|pmid=23614365}}</ref> |
|||
Recurrence biomarkers are used to predict if cancer is likely to come back after treatment. An example is the Oncotype DX® breast cancer assay.<ref name="NAME"/><ref>http://www.oncotypedx.com/HealthcareProfessional/Overview.aspx.</ref> This assay looks at several genes within a breast tumor sample and quantitatively indicates the probability that the patient’s cancer will return.<ref>http://www.oncotypedx.com/en-US/Breast/PatientCaregiver/OncoOverview.aspx</ref> |
|||
==Sensitivity and validity issues== |
==Sensitivity and validity issues== |
||
Revision as of 10:20, 26 April 2013
This article currently links to a large number of disambiguation pages (or back to itself). (April 2013) |
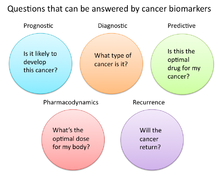
A cancer biomarker refers to a substance or process that is indicative of the presence of cancer in the body. A biomarker may be a molecule secreted by a tumor or a specific response of the body to the presence of cancer. Genetic, epigenetic, proteomic, glycomic, and imaging biomarkers can be used for cancer diagnosis, prognosis, and epidemiology. Ideally, such biomarkers can be assayed in non-invasively collected biofluids like blood or serum. [1]
While numerous challenges exist in translating biomarker research into the clinical space; a number of gene and protein based biomarkers have already been approved for use in patient care; including, AFP (Liver Cancer), BCR-ABL (Chronic Myeloid Leukemia), BRCA1 / BRCA2 (Breast/Ovarian Cancer), BRAF V600E (Melanoma/Colorectal Cancer), CA-125 (Ovarian Cancer) , CA19.9 (Pancreatic Cancer), CEA (Colorectal Cancer), EGFR (Non-small-cell lung carcinoma), HER-2 (Breast Cancer), KIT (Gastrointestinal stromal tumor), PSA (Prostate Specific Antigen) (Prostate Cancer), S100 (Melanoma), and many others. [2][3][4][5][6][7][8][9][10][11]
Definitions of Cancer Biomarkers
Organizations and publications vary in their definition of biomarker. In many areas of medicine, biomarkers are limited to proteins identifiable or measurable in the blood or urine. However, the term is often used to cover any molecular, biochemical, physiological, or anatomical property that can be quantified or measured.
The National Cancer Institute (NCI), in particular, defines biomarker as a: “A biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, or of a condition or disease. A biomarker may be used to see how well the body responds to a treatment for a disease or condition. Also called molecular marker and signature molecule." [12]
In cancer research and medicine, biomarkers are used in three primary ways: [13]
- To help diagnose conditions, as in the case of identifying early stage cancers (Diagnostic)
- To forecast how aggressive a condition is, as in the case of determining a patient's ability to fare in the absence of treatment (Prognostic)
- To predict how well a patient will respond to treatment (Predictive)
Role of Biomarkers in Cancer Research and Medicine
Uses of Biomarkers in Cancer Medicine
Risk Assessment
Cancer biomarkers, particular those associated with genetic mutations or epigenetic alterations, often offer a quantitative way to determine when individuals are predisposed to particular types of cancers. Notable examples of potentially predictive cancer biomarkers include mutations on genes KRAS, p53, EGFR, erbB2 for colorectal, esophageal, liver, and pancreatic cancer; mutations of genes BRCA1 and BRCA2 for breast and ovarian cancer; abnormal methylation of tumor suppressor genes p16, CDKN2B, and p14ARF for brain cancer; hypermethylation of MYOD1, CDH1, and CDH13 for cervical cancer; and hypermethylation of p16, p14, and RB1, for oral cancer.[14]
Diagnosis
Cancer biomarkers can also be useful in establishing a specific diagnosis. This is particularly the case when there is a need to determine whether tumors are of primary or metastatic origin. To make this distinction, researchers can screen the chromosomal alterations found on cells located in the primary tumor site against those found in the secondary site. If the alterations match, the secondary tumor can be identified as metastatic; whereas if the alterations differ, the secondary tumor can be identified as a distinct primary tumor.[15]
Prognosis and Treatment Predictions
Another use of biomarkers in cancer medicine is for disease prognosis, which take place after an individual has been diagnosed with cancer. Here biomarkers can be useful in determining the aggressiveness of an identified cancer as well as its likelihood of responding to a given treatment. In part, this is because tumors exhibiting particular biomarkers may be responsive to treatments tied to that biomarker's expression or presence. Examples of such prognostic biomarkers include elevated levels of metallopeptidase inhibitor 1 (TIMP1), a marker associated with more aggressive forms of multiple myeloma[16], elevated estrogen receptor (ER) and/or progesterone receptor (PR) expression, markers associated with better overall survival in patients with breast cancer[17][18]; HER2/neu gene amplification, a marker indicating a breast cancer will likely respond to trastuzumab treatment[19][20]; a mutation in exon 11 of the proto-oncogene c-KIT, indicating a gastrointestinal stromal tumor (GIST) will likely respond to imatinib treatment[21][22]; and mutations in the tyrosine kinase domain of EGFR1, indicating a patient's non-small-cell lung carcinoma (NSCLC) will likely respond to gefitinib or erlotinib treatment.[23][24]
Pharmacodynamics and Pharmacokinetics
Cancer biomarkers can also be used to determine the most effective treatment regime for a particular person's cancer.[25] Because of differences in each person's genetic makeup, some people metabolize or change the chemical structure of drugs differently. In some cases, decreased metabolism of certain drugs cancreate dangerous conditions in which high levels of the drug accumulate in the body. As such, drug dosing decisions in particular cancer treatments can benefit from screening for such biomarkers. An example is the gene encoding the enzyme thiopurine methyl-transferase (TPMPT).[26] Individuals with mutations in the TPMT gene are unable to metabolize large amounts of the leukemia drug, mercaptopurine, which potentially causes a fatal drop in white blood count for such patients. Patients with TPMT mutations are thus recommended to be given a lower dose of mercaptopurine for safety considerations. [27]
Monitoring Treatment Response
Cancer biomarkers have also shown utility in monitoring how well a treatemnt is working over time. Much research is going into this particular area, as successful biomarkers have the potential of providing significant cost reduction in patient care, as the current image-based tests such as CT and MRI for monitoring tumor status are highly costly.[28]
One notable biomarker garnering significant attention is the protein biomarker S100-beta in monitoring the response of malignant melanoma. In such melanomas, melanocytes, the cells that make pigment in our skin, produce the protein S100-beta in high concentrations dependent on the number of cancer cells. Response to treatment is asssociated levels of S100-beta in the blood of such individuals.[29][30] Additional laboratory research has shown that tumor cells undergoing apoptosis can release cellular components such as cytochrome c, nucleosomes, cleaved cytokeratin-18, and E-cadherin. Studies have found that these macromolecules and others can be found in circulation during cancer therapy, providing a potential source of clinical metrics for monitoring treatment.[28]
Recurrence
Lastly, cancer biomarkers offer value in predicting or monitoring cancer recurrence. The Oncotype DX® breast cancer assay is one such test used to predict the liklihood of breast cancer recurrence. Thist test is intended for women with early-stage (Stage I or II), node-negative, estrogen receptor-positive (ER+) invasive breast cancer who will be treated with hormone therapy. Oncotype DX looks at a panel of 21 genes in cells taken during tumor biopsy. The results of the test are given in the form of a recurrence score that indicates liklihood of recurrence at 10 years.[31] [32]
Sensitivity and validity issues
Nothing is ever perfect, and cancer biomarkers also abide by this guideline. The sensitivity for a cancer biomarker is often debated because its reliability varies with the sensitivity of the biomarker. For example, if detection of lung cancer biomarker X could signify that ALL people with detectable levels of X would get lung cancer, but people without X would not develop lung cancer, then biomarker X would be the optimal lung cancer predictor. But in reality, there are going to be some false positives (which tell healthy people they will get lung cancer) and false negatives (which tell at-risk people they will not develop cancer). However, the markers with high sensitivity and accuracy would be key in early cancer prevention or detection. Moreover, the optimal cancer biomarker is one that can be easily accessed from the body (i.e. blood, urine, tissue from biopsy).
Types of cancer biomarkers
Molecular cancer biomarkers
| Tumor Type | Biomarker |
| Breast | ER (estrogen receptor)[33] [34] |
| HER-2/neu [33] [34] | |
| colorectal | EGFR [33] [34] |
| KRAS [33] [35] | |
| UGT1A1 [33] [35] | |
| Gastric | HER-2/neu [33] |
| GIST | c-KIT [33] [36] |
| Leukemia/Lymphoma | CD20 Antigen [33] [37] |
| CD30 [33] [38] | |
| FIP1L1-PDGRFalpha [33] [39] | |
| PDGFR [33] [40] | |
| Philadelphia Chromosome (BCR/ABL) [33] [41] [42] | |
| PML/RAR alpha [33] [43] | |
| TPMT [33] [44] | |
| UGT1A1 [33] [45] | |
| Lung | ALK [33] [46] [47] |
| EGFR [33] [34] | |
| KRAS [33] [34] | |
| Melanoma | BRAF [33] [47] |
Other Examples of Biomarkers:
- Tumor Suppressors Lost in Cancer
- RNA
- Proteins found in body fluids or tissue.
- Examples: Prostate-specific antigen, and CA-125
Imaging techniques
- Examples: MRI, Mammography [49]
- Imaging disease biomarkers by magnetic resonance imaging (MRI)
MRI has the advantages of having very high spatial resolution and is very adept at morphological imaging and functional imaging. MRI does have several disadvantages though. First, MRI has a sensitivity of around 10−3 mol/L to 10−5 mol/L which, compared to other types of imaging, can be very limiting. This problem stems from the fact that the difference between atoms in the high energy state and the low energy state is very small. For example, at 1.5 tesla, a typical field strength for clinical MRI, the difference between high and low energy states is approximately 9 molecules per 2 million. Improvements to increase MR sensitivity include increasing magnetic field strength, and hyperpolarization via optical pumping or dynamic nuclear polarization. There are also a variety of signal amplification schemes based on chemical exchange that increase sensitivity.
To achieve molecular imaging of disease biomarkers using MRI, targeted MRI contrast agents with high specificity and high relaxivity (sensitivity) are required. To date, many studies have been devoted to developing targeted-MRI contrast agents to achieve molecular imaging by MRI. Commonly, peptides, antibodies, or small ligands, and small protein domains, such as HER-2 affibodies, have been applied to achieve targeting. To enhance the sensitivity of the contrast agents, these targeting moieties are usually linked to high payload MRI contrast agents or MRI contrast agents with high relaxivities.[50]
References
- ^ Mishra, Alok (2010). "Cancer Biomarkers: Are We Ready for the Prime Time?". Cancers. 2 (1): 190-208.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Rhea, Jeanne (2011). "Cancer Biomarkers: Surviving the journey from bench to bedside". Medical Laboratory Observer. Retrieved 26 April 2013.
{{cite web}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Behne, Tara (1 January 2012). "Biomarkers for Hepatocellular Carcinoma". International Journal of Hepatology. 2012: 1–7. doi:10.1155/2012/859076.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: date and year (link) CS1 maint: unflagged free DOI (link) - ^ Musolino, A (2007 Jun). "BRCA mutations, molecular markers, and clinical variables in early-onset breast cancer: a population-based study". Breast (Edinburgh, Scotland). 16 (3): 280–92. PMID 17257844.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Dienstmann, R (2011 Mar). "BRAF as a target for cancer therapy". Anti-cancer agents in medicinal chemistry. 11 (3): 285–95. PMID 21426297.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Lamparella, N (2013). "Impact of genetic markers on treatment of non-small cell lung cancer". Advances in experimental medicine and biology. 779: 145–64. PMID 23288638.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Orphanos, G (2012). "Targeting the HER2 receptor in metastatic breast cancer". Hematology/oncology and stem cell therapy. 5 (3): 127–37. PMID 23095788.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DePrimo, S. E. (8 September 2009). "Circulating Levels of Soluble KIT Serve as a Biomarker for Clinical Outcome in Gastrointestinal Stromal Tumor Patients Receiving Sunitinib following Imatinib Failure". Clinical Cancer Research. 15 (18): 5869–5877. doi:10.1158/1078-0432.CCR-08-2480.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Bantis, A (2012 Sep-Dec). "Prostatic specific antigen and bone scan in the diagnosis and follow-up of prostate cancer. Can diagnostic significance of PSA be increased?". Hellenic journal of nuclear medicine. 15 (3): 241–6. PMID 23227460.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Kruijff, S (2012 Apr). "The current status of S-100B as a biomarker in melanoma". European journal of surgical oncology : the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 38 (4): 281–5. PMID 22240030.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Ludwig, JA (2005 Nov). "Biomarkers in cancer staging, prognosis and treatment selection". Nature reviews. Cancer. 5 (11): 845–56. PMID 16239904.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ "biomarker". NCI Dictionary of Cancer Terms. National Cancer Institute.
- ^ "Biomarkers in Cancer: An Introductory Guide for Advocates" (PDF). Research Advocay Network. 2010. Retrieved 26 April 2013.
- ^ Verma, M (2006 Oct). "Genetic and epigenetic biomarkers in cancer diagnosis and identifying high risk populations". Critical reviews in oncology/hematology. 60 (1): 9–18. PMID 16829121.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Leong, PP (1998 Jul 1). "Distinguishing second primary tumors from lung metastases in patients with head and neck squamous cell carcinoma". Journal of the National Cancer Institute. 90 (13): 972–7. PMID 9665144.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Terpos E, Dimopoulos MA, Shrivastava V; et al. (2010). "High levels of serum TIMP-1 correlate with advanced disease and predict for poor survival in patients with multiple myeloma treated with novel agents". Leuk. Res. 34 (3): 399–402. doi:10.1016/j.leukres.2009.08.035. PMID 19781774.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Kuukasjärvi, T (1996 Sep). "Loss of estrogen receptor in recurrent breast cancer is associated with poor response to endocrine therapy". Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 14 (9): 2584–9. PMID 8823339.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Harris, L (2007 Nov 20). "American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer". Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 25 (33): 5287–312. PMID 17954709.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Kröger, N (2006 Jan 1). "Prognostic and predictive effects of immunohistochemical factors in high-risk primary breast cancer patients". Clinical cancer research : an official journal of the American Association for Cancer Research. 12 (1): 159–68. PMID 16397038.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Vrbic, S (2013 Jan-Mar). "Current and future anti-HER2 therapy in breast cancer". Journal of B.U.ON. : official journal of the Balkan Union of Oncology. 18 (1): 4–16. PMID 23613383.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Yoo, C (2013 Apr 17). "Efficacy, safety, and pharmacokinetics of imatinib dose escalation to 800 mg/day in patients with advanced gastrointestinal stromal tumors". Investigational new drugs. PMID 23591629.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help); no-break space character in|title=at position 74 (help) - ^ Demetri, GD (2006 Oct 14). "Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial". Lancet. 368 (9544): 1329–38. PMID 17046465.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Herbst, RS (2005 Sep 1). "TRIBUTE: a phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer". Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 23 (25): 5892–9. PMID 16043829.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Lynch, TJ (2004 May 20). "Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib". The New England journal of medicine. 350 (21): 2129–39. PMID 15118073.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Sawyers CL (2008). "The cancer biomarker problem". Nature. 452 (7187): 548–52. doi:10.1038/nature06913. PMID 18385728.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Cite error: The named reference
NAMEwas invoked but never defined (see the help page). - ^ Relling MV, Hancock ML, Rivera GK; et al. (1999). "Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus". J. Natl. Cancer Inst. 91 (23): 2001–8. PMID 10580024.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Schneider, John (2012). "Economics of Cancer Biomarkers". Personalized Medicine. 9 (8): 829-837.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Henze, G (1997). "Serum S100--a marker for disease monitoring in metastatic melanoma". Dermatology (Basel, Switzerland). 194 (3): 208–12. PMID 9187834.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Harpio, R (2004 Jul). "S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma". Clinical biochemistry. 37 (7): 512–8. PMID 15234232.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Lamond, NW (2013 Apr). "Is the 21-gene recurrence score a cost-effective assay in endocrine-sensitive node-negative breast cancer?". Expert review of pharmacoeconomics & outcomes research. 13 (2): 243–50. PMID 23570435.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Biroschak, JR (2013 May). "Impact of Oncotype DX on Treatment Decisions in ER-Positive, Node-Negative Breast Cancer with Histologic Correlation". The breast journal. 19 (3): 269–75. PMID 23614365.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d e f g h i j k l m n o p q r s "Table of Pharmacogenomic Biomarkers in Drug Labels". U.S Food and Drug Administration.
- ^ a b c d e "Tumor Markers Fact Sheet" (PDF). American Cancer Society.
- ^ a b Lenz, Heinz-Josef. EdBk.GI.Colo.04.pdf "Established Biomarkers in Colon Cancer" (PDF). American Society of Clinical Oncology’s 2009 Educational Book.
{{cite web}}: Check|url=value (help) - ^ Gonzalez RS, Carlson G, Page AJ, Cohen C (2011). "Gastrointestinal stromal tumor markers in cutaneous melanomas: relationship to prognostic factors and outcome". Am. J. Clin. Pathol. 136 (1): 74–80. doi:10.1309/AJCP9KHD7DCHWLMO. PMID 21685034.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Tam CS, Otero-Palacios J, Abruzzo LV; et al. (2008). "Chronic lymphocytic leukaemia CD20 expression is dependent on the genetic subtype: a study of quantitative flow cytometry and fluorescent in-situ hybridization in 510 patients". Br. J. Haematol. 141 (1): 36–40. doi:10.1111/j.1365-2141.2008.07012.x. PMID 18324964.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Zhang M, Yao Z, Patel H; et al. (2007). "Effective therapy of murine models of human leukemia and lymphoma with radiolabeled anti-CD30 antibody, HeFi-1". Proc. Natl. Acad. Sci. U.S.A. 104 (20): 8444–8. doi:10.1073/pnas.0702496104. PMC 1895969. PMID 17488826.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Yamada Y, Sanchez-Aguilera A, Brandt EB; et al. (2008). "FIP1L1/PDGFRalpha synergizes with SCF to induce systemic mastocytosis in a murine model of chronic eosinophilic leukemia/hypereosinophilic syndrome". Blood. 112 (6): 2500–7. doi:10.1182/blood-2007-11-126268. PMID 18539901.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Nimer SD (2008). "Myelodysplastic syndromes". Blood. 111 (10): 4841–51. doi:10.1182/blood-2007-08-078139. PMID 18467609.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Ottmann O, Dombret H, Martinelli G; et al. (2007). "Dasatinib induces rapid hematologic and cytogenetic responses in adult patients with Philadelphia chromosome positive acute lymphoblastic leukemia with resistance or intolerance to imatinib: interim results of a phase 2 study". Blood. 110 (7): 2309–15. doi:10.1182/blood-2007-02-073528. PMID 17496201.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Boulos N, Mulder HL, Calabrese CR; et al. (2011). "Chemotherapeutic agents circumvent emergence of dasatinib-resistant BCR-ABL kinase mutations in a precise mouse model of Philadelphia chromosome-positive acute lymphoblastic leukemia". Blood. 117 (13): 3585–95. doi:10.1182/blood-2010-08-301267. PMC 3072880. PMID 21263154.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ O'Connell PA, Madureira PA, Berman JN, Liwski RS, Waisman DM (2011). "Regulation of S100A10 by the PML-RAR-α oncoprotein". Blood. 117 (15): 4095–105. doi:10.1182/blood-2010-07-298851. PMID 21310922.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Duffy MJ, Crown J (2008). "A personalized approach to cancer treatment: how biomarkers can help". Clin. Chem. 54 (11): 1770–9. doi:10.1373/clinchem.2008.110056. PMID 18801934.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Ribrag V, Koscielny S, Casasnovas O; et al. (2009). "Pharmacogenetic study in Hodgkin lymphomas reveals the impact of UGT1A1 polymorphisms on patient prognosis". Blood. 113 (14): 3307–13. doi:10.1182/blood-2008-03-148874. PMID 18768784.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Li Y, Ye X, Liu J, Zha J, Pei L (2011). "Evaluation of EML4-ALK fusion proteins in non-small cell lung cancer using small molecule inhibitors". Neoplasia. 13 (1): 1–11. PMC 3022423. PMID 21245935.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Pao W, Girard N (2011). "New driver mutations in non-small-cell lung cancer". Lancet Oncol. 12 (2): 175–80. doi:10.1016/S1470-2045(10)70087-5. PMID 21277552.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Bartels CL, Tsongalis GJ (2009). "MicroRNAs: novel biomarkers for human cancer". Clin. Chem. 55 (4): 623–31. doi:10.1373/clinchem.2008.112805. PMID 19246618.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Dalton WS, Friend SH (2006). "Cancer biomarkers—an invitation to the table". Science. 312 (5777): 1165–8. doi:10.1126/science.1125948. PMID 16728629.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Shenghui, Xue (17). "Design of a novel class of protein-based magnetic resonance imaging contrast agents for the molecular imaging of cancer biomarkers". Wiley Interdiscip Rev Nanomed Nanobiotechnol. 5 (2). doi:10.1002/wnan.1205. PMID 23335551.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help)
External links
- CancerDriver: open database of cancer biomarkers with links to references and recruiting clinical trials.
- The latest news about oncology biomarkers
