Coronavirus envelope protein
| Envelope protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
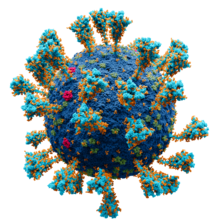 Model of the external structure of the SARS-CoV-2 virion.[1]
● Blue: envelope ● Turquoise: spike glycoprotein (S) ● Red: envelope proteins (E) ● Green: membrane proteins (M) ● Orange: glycans | |||||||||
| Identifiers | |||||||||
| Symbol | CoV_E | ||||||||
| Pfam | PF02723 | ||||||||
| InterPro | IPR003873 | ||||||||
| PROSITE | PS51926 | ||||||||
| |||||||||
The envelope (E) protein is the smallest and least well-characterized of the four major structural proteins found in coronavirus virions.[2][3][4] It is an integral membrane protein less than 110 amino acid residues long;[2] in SARS-CoV-2, the causative agent of Covid-19, the E protein is 75 residues long.[5] Although it is not necessarily essential for viral replication, absence of the E protein may produce abnormally assembled viral capsids or reduced replication.[2][3] E is a multifunctional protein[6] and, in addition to its role as a structural protein in the viral capsid, it is thought to be involved in viral assembly, likely functions as a viroporin, and is involved in viral pathogenesis.[2][5]
Structure
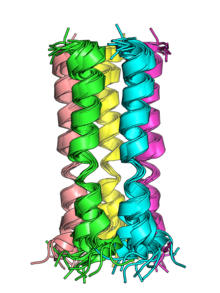
The E protein consists of a short hydrophilic N-terminal region, a hydrophobic helical transmembrane domain, and a somewhat hydrophilic C-terminal region. In SARS-CoV and SARS-CoV-2, the C-terminal region contains a PDZ domain binding motif (PBM).[2][5] This feature appears to be conserved only in the alpha and beta coronavirus groups, but not gamma.[2] In the beta and gamma groups, a conserved proline residue is found in the C-terminal region likely involved in targeting the protein to the Golgi.[2]
The transmembrane helices of the E proteins of SARS-CoV and SARS-CoV-2 can oligomerize and have been shown in vitro to form pentameric structures with central pores that serve as cation-selective ion channels.[5] Both viruses' E protein pentamers have been structurally characterized by nuclear magnetic resonance spectroscopy.[5][7]
The membrane topology of the E protein has been studied in a number of coronaviruses with inconsistent results; the protein's orientation in the membrane may be variable.[3] The balance of evidence suggests the most common orientation has the C-terminus oriented toward the cytoplasm.[8] Studies of SARS-CoV-2 E protein are consistent with this orientation.[5][9]
Post-translational modifications
In some, but not all, coronaviruses, the E protein is post-translationally modified by palmitoylation on conserved cysteine residues.[2][8] In the SARS-CoV E protein, one glycosylation site has been observed, which may influence membrane topology;[8] however, the functional significance of E glycosylation is unclear.[2] Ubiquitination of SARS-CoV E has also been described, though its functional significance is also not known.[2]
Expression and localization
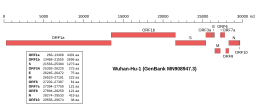 Genomic organisation of isolate Wuhan-Hu-1, the earliest sequenced sample of SARS-CoV-2, indicating the location of the E gene | |
| NCBI genome ID | 86693 |
|---|---|
| Genome size | 29,903 bases |
| Year of completion | 2020 |
| Genome browser (UCSC) | |
The E protein is expressed at high abundance in infected cells. However, only a small amount of the total E protein produced is found in assembled virions.[2][4] E protein is localized to the endoplasmic reticulum, Golgi apparatus, and endoplasmic-reticulum–Golgi intermediate compartment (ERGIC), the intracellular compartment that gives rise to the coronavirus viral envelope.[2][5]
Function
Essentiality
Studies in different coronaviruses have reached different conclusions about whether E is essential to viral replication. In some coronaviruses, including MERS-CoV, E has been reported to be essential.[10] In others, including mouse coronavirus[11] and SARS-CoV, E is not essential, though its absence reduces viral titer,[12] in some cases by introducing propagation defects or causing abnormal capsid morphology.[2]
Virions and viral assembly

The E protein is found in assembled virions where it forms protein-protein interactions with the coronavirus membrane protein (M), the most abundant of the four structural proteins contained in the viral capsid.[2][4] The interaction between E and M occurs through their respective C-termini on the cytoplasmic side of the membrane.[2] In most coronaviruses, E and M are sufficient to form virus-like particles,[2][4] though SARS-CoV has been reported to depend on N as well.[14] There is good evidence that E is involved in inducing membrane curvature to create the typical spherical coronavirus virion.[2][15] It is likely that E is involved in viral budding or scission, although its role in this process has not been well characterized.[2][4][15]
Viroporin
In its pentameric state, E forms cation-selective ion channels and likely functions as a viroporin.[5] This may disrupt ion homeostasis, alter membrane permeability, and modulate pH in the host cell, which may facilitate viral release.[2][4] The E protein's role as a viroporin appears to be involved in pathogenesis and may be related to activation of the inflammasome.[3][16] In SARS-CoV, mutations that disrupt E's ion channel function result in attenuated pathogenesis in animal models despite little effect on viral growth.[10]
Interactions with host proteins

Protein-protein interactions between E and proteins in the host cell are best described in SARS-CoV and occur via the C-terminal PDZ domain binding motif. The SARS-CoV E protein has been reported to interact with five host cell proteins: Bcl-xL, PALS1, syntenin, sodium/potassium (Na+/K+) ATPase α-1 subunit, and stomatin.[2] The interaction with PALS1 may be related to pathogenesis via the resulting disruption in tight junctions.[3][10] This interaction has also been identified in SARS-CoV-2.[17]
Evolution and conservation
The sequence of the E protein is not well conserved across coronavirus genera, with sequence identities reaching under 30%.[12] In laboratory experiments on mouse hepatitis virus, substitution of E proteins from different coronaviruses, even from different groups, could produce viable viruses, suggesting that significant sequence diversity can be tolerated in functional E proteins.[18] The SARS-CoV-2 E protein is very similar to that of SARS-CoV, with three substitutions and one deletion.[4] A study of SARS-CoV-2 sequences suggests that the E protein is evolving relatively slowly compared to other structural proteins.[19] The conserved nature of the envelope protein among SARS-CoV and SARS-CoV-2 variants has led it to be researched as a potential target for universal coronavirus vaccine development.[20][21]
References
- ^ Solodovnikov, Alexey; Arkhipova, Valeria (2021-07-29). "Достоверно красиво: как мы сделали 3D-модель SARS-CoV-2" [Truly beautiful: how we made the SARS-CoV-2 3D model] (in Russian). N+1. Archived from the original on 2021-07-30. Retrieved 30 July 2021.
- ^ a b c d e f g h i j k l m n o p q r s t Schoeman, Dewald; Fielding, Burtram C. (December 2019). "Coronavirus envelope protein: current knowledge". Virology Journal. 16 (1): 69. doi:10.1186/s12985-019-1182-0. PMC 6537279. PMID 31133031.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e Schoeman, Dewald; Fielding, Burtram C. (2020-09-03). "Is There a Link Between the Pathogenic Human Coronavirus Envelope Protein and Immunopathology? A Review of the Literature". Frontiers in Microbiology. 11: 2086. doi:10.3389/fmicb.2020.02086. PMC 7496634. PMID 33013759.
- ^ a b c d e f g h Cao, Yipeng; Yang, Rui; Lee, Imshik; Zhang, Wenwen; Sun, Jiana; Wang, Wei; Meng, Xiangfei (June 2021). "Characterization of the SARS‐CoV ‐2 E Protein: Sequence, Structure, Viroporin, and Inhibitors". Protein Science. 30 (6): 1114–1130. doi:10.1002/pro.4075. ISSN 0961-8368. PMC 8138525. PMID 33813796.
- ^ a b c d e f g h i Mandala, Venkata S.; McKay, Matthew J.; Shcherbakov, Alexander A.; Dregni, Aurelio J.; Kolocouris, Antonios; Hong, Mei (December 2020). "Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers". Nature Structural & Molecular Biology. 27 (12): 1202–1208. doi:10.1038/s41594-020-00536-8. PMC 7718435. PMID 33177698.
- ^ Liu, D. X.; Yuan, Q.; Liao, Y. (August 2007). "Coronavirus envelope protein: A small membrane protein with multiple functions". Cellular and Molecular Life Sciences. 64 (16): 2043–2048. doi:10.1007/s00018-007-7103-1. PMC 7079843. PMID 17530462.
- ^ Surya, Wahyu; Li, Yan; Torres, Jaume (June 2018). "Structural model of the SARS coronavirus E channel in LMPG micelles". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1860 (6): 1309–1317. doi:10.1016/j.bbamem.2018.02.017. PMC 7094280. PMID 29474890.
- ^ a b c Fung, To Sing; Liu, Ding Xiang (June 2018). "Post-translational modifications of coronavirus proteins: roles and function". Future Virology. 13 (6): 405–430. doi:10.2217/fvl-2018-0008. PMC 7080180. PMID 32201497.
- ^ Duart, Gerard; García-Murria, Mª Jesús; Grau, Brayan; Acosta-Cáceres, José M.; Martínez-Gil, Luis; Mingarro, Ismael (September 2020). "SARS-CoV-2 envelope protein topology in eukaryotic membranes". Open Biology. 10 (9): 200209. doi:10.1098/rsob.200209. PMC 7536074. PMID 32898469.
- ^ a b c DeDiego, Marta L.; Nieto-Torres, Jose L.; Jimenez-Guardeño, Jose M.; Regla-Nava, Jose A.; Castaño-Rodriguez, Carlos; Fernandez-Delgado, Raul; Usera, Fernando; Enjuanes, Luis (December 2014). "Coronavirus virulence genes with main focus on SARS-CoV envelope gene". Virus Research. 194: 124–137. doi:10.1016/j.virusres.2014.07.024. PMC 4261026. PMID 25093995.
- ^ Kuo, Lili; Masters, Paul S. (2003-04-15). "The Small Envelope Protein E Is Not Essential for Murine Coronavirus Replication". Journal of Virology. 77 (8): 4597–4608. doi:10.1128/JVI.77.8.4597-4608.2003. PMC 152126. PMID 12663766.
- ^ a b Ruch, Travis R.; Machamer, Carolyn E. (2012-03-08). "The Coronavirus E Protein: Assembly and Beyond". Viruses. 4 (3): 363–382. doi:10.3390/v4030363. PMC 3347032. PMID 22590676.
- ^ Goodsell, David S.; Voigt, Maria; Zardecki, Christine; Burley, Stephen K. (6 August 2020). "Integrative illustration for coronavirus outreach". PLOS Biology. 18 (8): e3000815. doi:10.1371/journal.pbio.3000815. PMC 7433897. PMID 32760062.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Siu, Y. L.; Teoh, K. T.; Lo, J.; Chan, C. M.; Kien, F.; Escriou, N.; Tsao, S. W.; Nicholls, J. M.; Altmeyer, R.; Peiris, J. S. M.; Bruzzone, R.; Nal, B. (15 November 2008). "The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-Like Particles". Journal of Virology. 82 (22): 11318–11330. doi:10.1128/JVI.01052-08. PMC 2573274. PMID 18753196.
- ^ a b Alsaadi, Entedar A J; Jones, Ian M (April 2019). "Membrane binding proteins of coronaviruses". Future Virology. 14 (4): 275–286. doi:10.2217/fvl-2018-0144. PMC 7079996. PMID 32201500.
- ^ Nieto-Torres, Jose L.; DeDiego, Marta L.; Verdiá-Báguena, Carmina; Jimenez-Guardeño, Jose M.; Regla-Nava, Jose A.; Fernandez-Delgado, Raul; Castaño-Rodriguez, Carlos; Alcaraz, Antonio; Torres, Jaume; Aguilella, Vicente M.; Enjuanes, Luis (1 May 2014). "Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis". PLOS Pathogens. 10 (5): e1004077. doi:10.1371/journal.ppat.1004077. PMC 4006877. PMID 24788150.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Chai, Jin; Cai, Yuanheng; Pang, Changxu; Wang, Liguo; McSweeney, Sean; Shanklin, John; Liu, Qun (December 2021). "Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1". Nature Communications. 12 (1): 3433. Bibcode:2021NatCo..12.3433C. doi:10.1038/s41467-021-23533-x. PMC 8187709. PMID 34103506.
- ^ Kuo, Lili; Hurst, Kelley R.; Masters, Paul S. (March 2007). "Exceptional Flexibility in the Sequence Requirements for Coronavirus Small Envelope Protein Function". Journal of Virology. 81 (5): 2249–2262. doi:10.1128/JVI.01577-06. PMC 1865940. PMID 17182690.
- ^ Rahman, M. Shaminur; Hoque, M. Nazmul; Islam, M. Rafiul; Islam, Israt; Mishu, Israt Dilruba; Rahaman, Md. Mizanur; Sultana, Munawar; Hossain, M. Anwar (March 2021). "Mutational insights into the envelope protein of SARS-CoV-2". Gene Reports. 22: 100997. doi:10.1016/j.genrep.2020.100997. PMC 7723457. PMID 33319124.
- ^ Bhattacharya, Shreya; Banerjee, Arundhati; Ray, Sujay (2021-03-01). "Development of new vaccine target against SARS-CoV2 using envelope (E) protein: An evolutionary, molecular modeling and docking based study". International Journal of Biological Macromolecules. 172: 74–81. doi:10.1016/j.ijbiomac.2020.12.192. PMC 7833863. PMID 33385461.
- ^ Chen, Jinni; Deng, Yao; Huang, Baoying; Han, Di; Wang, Wen; Huang, Mengjing; Zhai, Chengcheng; Zhao, Zhimin; Yang, Ren; Zhao, Ying; Wang, Wenling; Zhai, Desheng; Tan, Wenjie (2022-02-24). "DNA Vaccines Expressing the Envelope and Membrane Proteins Provide Partial Protection Against SARS-CoV-2 in Mice". Frontiers in Immunology. 13: 827605. doi:10.3389/fimmu.2022.827605. ISSN 1664-3224. PMC 8907653. PMID 35281016.

