User talk:Boghog: Difference between revisions
→IUPHAR links: great! |
|||
| Line 129: | Line 129: | ||
: Hi Alexander. It is great to have some experts like yourself contributing to these Gene Wiki articles! Concerning [[Calcium-activated potassium channel subunit alpha-1]], I have no objection renaming this article to [[KCNMA1]]. I am not so sure about merging the [[BK channel]] article into [[KCNMA1]] however. The later is a gene/protein specific article while the former is about the alpha tetramer + optional beta subunits. I think it makes some sense to have one parent article that covers both alpha and beta subunits ([[BK channel]]) plus separate gene/protein specific articles ([[KCNMA1]], [[KCNMB1]], [[KCNMB2]], [[KCNMB3]], [[KCNMB4]]). One thing that probably should be moved is the {{tl|Infobox protein family}} (Pfam calcium-activated BK potassium channel alpha subunit) from [[BK channel]] to [[KCNMA1]]. Thoughts? [[User:Boghog|Boghog]] ([[User talk:Boghog#top|talk]]) 18:37, 11 April 2012 (UTC) |
: Hi Alexander. It is great to have some experts like yourself contributing to these Gene Wiki articles! Concerning [[Calcium-activated potassium channel subunit alpha-1]], I have no objection renaming this article to [[KCNMA1]]. I am not so sure about merging the [[BK channel]] article into [[KCNMA1]] however. The later is a gene/protein specific article while the former is about the alpha tetramer + optional beta subunits. I think it makes some sense to have one parent article that covers both alpha and beta subunits ([[BK channel]]) plus separate gene/protein specific articles ([[KCNMA1]], [[KCNMB1]], [[KCNMB2]], [[KCNMB3]], [[KCNMB4]]). One thing that probably should be moved is the {{tl|Infobox protein family}} (Pfam calcium-activated BK potassium channel alpha subunit) from [[BK channel]] to [[KCNMA1]]. Thoughts? [[User:Boghog|Boghog]] ([[User talk:Boghog#top|talk]]) 18:37, 11 April 2012 (UTC) |
||
:: I had the same thought initially, but actually the entire BK article is only relevant to the alpha subunit and doesn't say anything substantial about the beta subunits. I was also persuaded by the fact that other articles and templates already provide overview of the complex and list the beta subunits: [[Calcium-activated_potassium_channel]] and [[Template:Ion_channels]]. [[User:AlexanderPico|AlexanderPico]] ([[User talk:AlexanderPico|talk]]) 19:22, 11 April 2012 (UTC) |
|||
Revision as of 19:22, 11 April 2012
|
This page has archives. Sections older than 30 days may be automatically archived by Lowercase sigmabot III when more than 4 sections are present. |
PDB links in drug box
| vemurafenib | |
|---|---|
| Drug mechanism | |
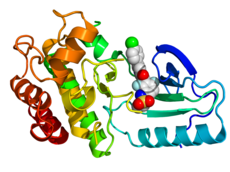 Crystallographic structure of B-Raf (rainbow colored, N-terminus = blue, C-terminus = red) complexed with vemurafenib (spheres, carbon = white, oxygen = red, nitrogen = blue, chlorine = green, fluorine = cyan, sulfur = yellow).[1] | |
| Therapeutic use | melanoma |
| Biological target | BRAF |
| Mechanism of action | protein kinase inhibitor |
| External links | |
| ATC code | L01XE15 |
| PDB ligand id | 032: PDBe, RCSB PDB |
| LIGPLOT | 3og7 |
Hi. I see that you have some edits to drugbox in sandbox2. I was wondering if we could add links to PDB entries with molecules. There is no information in these boxes on "bound structure". I was wondering if the following kind of link woul dbe useful to add to this box - e.g. for Nicotine I have added http://www.ebi.ac.uk/pdbe-srv/PDBeXplore/ligand/?ligand=NCT under external links. I can easily provide mapping between PDB three letter codes and InCHI string or keys or mapping to ChEMBL id's. A2-25 (talk) 20:42, 25 February 2012 (UTC)
- Yes, this certainly could be done and I would personally support it. My only reservation is that the {{drugbox}} is becoming incredibly complex and we must be careful of feature creep. Another issue is where to place the link. Incredibly this infobox does not currently contain any information about target or mechanism other than the ATC code. Perhaps we should add a mechanism section (perhaps called "pharmacodynamic data") that includes the ATC code, a wiki link to the biological target, and other basic information (whether the drug acts as agonist, antagonist, irreversible inhibitor, etc.). The adding the PDB link to this section would make the most sense. There are a lot of editors who have strong opinions about this template, so we need to build consensus before making any changes. Boghog (talk) 21:59, 25 February 2012 (UTC)
- On second thought, it might be better to create a new infobox containing drug mechanism information. I have created a prototype {{infobox drug mechanism}} template and a filled out example can be seen to the right. The advantage of having a separate infobox is that it can be placed in the mechanism of action section of the drug article and in addition, we can add a image of the drug/protein complex. Thoughts? Boghog (talk) 06:43, 26 February 2012 (UTC)
- ^ PDB: 3OG7; Bollag G; Hirth P; et al. (2010). "Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma". Nature. 467 (7315): 596–599. doi:10.1038/nature09454. PMC 2948082. PMID 20823850.
{{cite journal}}: Unknown parameter|author-separator=ignored (help); Unknown parameter|month=ignored (help)
- This would be a better way of doing it. A new box woud allow us to add extra information later. e.g. I would very much like to start adding information about the binding sites etc. Have a look at - http://www.ebi.ac.uk/pdbe-srv/pdbexpress/ligandEnvironment/showEnvironment?ligand=RTL which gives more information on binding residues. Another such analysis that might be useful is - http://www.ebi.ac.uk/pdbe-srv/pdbexpress/EC/showEnzymes?ligand=MAN. We can also add extra information on targets if such information is easily accessible via link. So I support adding an extra box. One of the images below is ligand interaction image so we can add that as you suggest. Suggestions/thoughts? A2-25 (talk) 08:42, 26 February 2012 (UTC)
- While there are exceptions, most drugs bind with high affinity to a single biological target that in turn is responsible for the therapeutic effect. Hence for drugs, I think a link to a LigPlot or PoseView representation of the binding cavity (see for example LigPlot O32/3og7 or PoseView O32/3og7) would be more relevant. The links that you have provided above to composite binding site information may be of more relevant for promiscuous ligands that bind to many different types of proteins, and hence would be more appropriate to add for example to the {{chembox}} template in the manose article. Boghog (talk) 10:06, 26 February 2012 (UTC)
- This would be a better way of doing it. A new box woud allow us to add extra information later. e.g. I would very much like to start adding information about the binding sites etc. Have a look at - http://www.ebi.ac.uk/pdbe-srv/pdbexpress/ligandEnvironment/showEnvironment?ligand=RTL which gives more information on binding residues. Another such analysis that might be useful is - http://www.ebi.ac.uk/pdbe-srv/pdbexpress/EC/showEnzymes?ligand=MAN. We can also add extra information on targets if such information is easily accessible via link. So I support adding an extra box. One of the images below is ligand interaction image so we can add that as you suggest. Suggestions/thoughts? A2-25 (talk) 08:42, 26 February 2012 (UTC)
That sounds like a good plan. I don't think we should add two links Ligplot and poseView since both show same/very similar information and LigPlot is something people are more familier with so I prefer adding link to LigPlot but do not have any objection if you think both links are useful. Let me know what information you need to implement this. Another suggestion I have is to link the ligand id to something like http://pdbe.org/chem/032 so users can see PDB definition of the molecule with atom names etc. A2-25 (talk) 12:37, 26 February 2012 (UTC)
- Thanks for the suggestions. I agree Ligplot is more widely known and the PDBe ligand ID link that you mentioned is more appropriate. I have implemented both in the prototype template and in the example to the right. Is the LIgplot link OK? Boghog (talk) 14:24, 26 February 2012 (UTC)
I am close to getting the server to give best image picture. I wanted to know your opinion on adding extra picture in wikimedia. I am thinking of adding all images similar to ones on
http://www.ebi.ac.uk/pdbe-apps/widgets/PDBimages/cb/1s/index.html which are images for 1cbs or http://www.ebi.ac.uk/pdbe-apps/widgets/PDBimages/e9/14/index.html for 1e94. We can then think about using these in other pages e.g. showing Pfam domain on structures or CATH and SCOP domains. In any case all these images are freely available. Let me know your opinion A2-25 (talk) 20:42, 25 February 2012 (UTC)
- Great! Perhaps we should first concentrate on adding graphics to {{GNF Protein box}} templates that currently do not have graphics. For that purpose, a bot could download as needed graphics and upload them to Wikimedia commons. A tricker question is what to do about templates that already have graphics. Some of these were manually added and I would be very hesitant on replacing those. The remainder are high quality ray traced PyMol images that User:Emw worked very hard at producing. In some cases, these have small coverage of the UniProt sequence and better alternatives exist. These images should be replaced. However I would be reluctant to replace the remainder. Thoughts? Boghog (talk) 21:59, 25 February 2012 (UTC)
- I agree we will not use all the images immediately. I will still upload all the images so if we want image we do not have to wait for adding these images. We at EBI must have enough band width to do this. I am also taking help from other wiser wikipedia users to see how this can be done. If nothing comes off it then we anyway have a plan for uploading these images. I maintain the SIFTS (pdbe.org/sifts) project which keeps up-to-date information on links between PDB and all other databases. This information is used by most databases (RCSB, Pfam, CATH, SCOP to name a few) so I suppose I am in a position to give all information about mappings and any changes to those mappings. So in summary, I agree with you that we can not use all the images and replace old ones but then will it be useful to create a new box with title "representative structure" and add the image. We could even make it (I don't know if that is possible) such that we add all the extra images that show mappings to other databases mentioned above. suggestions/thoughts? A2-25 (talk) 08:42, 26 February 2012 (UTC)
- One needs to be careful about uploading large numbers of images to Wikimedia commons since images that are not used in articles tend to get deleted. It appears that you have already generated and stored these images at the EBI. Hence it might be a better solution to upload these images to Wikipedia only as needed. The extra links also are of interest, particularly Pfam. Since Wikipedia article already exist for many Pfam entries, the links should preferably take the form of internal Wiki links. Boghog (talk) 10:06, 26 February 2012 (UTC)
- No problem. As you have noticed all images are ready so we can add it as necessary. I wanted to add them since these are freely available so if someone is searching for a particular image they can find it easily. But if unused images get deleted it is not that useful to upload all images. As you suggest adding Pfam mapping might be useful so I can concentrate on uploading best structure images first and once we have that working then Pfam mapping images. I have no problem if these are internal wiki links. A2-25 (talk) 12:37, 26 February 2012 (UTC)
- Sounds good. Just to clarify, it would be useful to have the Pfam accession numbers so that the relevant Wikipedia Pfam articles can be identified and if the corresponding Wikipedia article has not yet been created, we can include an external link using the {{Pfam}} template. Boghog (talk) 14:24, 26 February 2012 (UTC)
- I am not sure I replied to this. Have a look at -
- These pages give all the mappings from PDB to other databases.A2-25 (talk) 15:06, 17 March 2012 (UTC)
- Sounds good. Just to clarify, it would be useful to have the Pfam accession numbers so that the relevant Wikipedia Pfam articles can be identified and if the corresponding Wikipedia article has not yet been created, we can include an external link using the {{Pfam}} template. Boghog (talk) 14:24, 26 February 2012 (UTC)
- No problem. As you have noticed all images are ready so we can add it as necessary. I wanted to add them since these are freely available so if someone is searching for a particular image they can find it easily. But if unused images get deleted it is not that useful to upload all images. As you suggest adding Pfam mapping might be useful so I can concentrate on uploading best structure images first and once we have that working then Pfam mapping images. I have no problem if these are internal wiki links. A2-25 (talk) 12:37, 26 February 2012 (UTC)
- One needs to be careful about uploading large numbers of images to Wikimedia commons since images that are not used in articles tend to get deleted. It appears that you have already generated and stored these images at the EBI. Hence it might be a better solution to upload these images to Wikipedia only as needed. The extra links also are of interest, particularly Pfam. Since Wikipedia article already exist for many Pfam entries, the links should preferably take the form of internal Wiki links. Boghog (talk) 10:06, 26 February 2012 (UTC)
- I agree we will not use all the images immediately. I will still upload all the images so if we want image we do not have to wait for adding these images. We at EBI must have enough band width to do this. I am also taking help from other wiser wikipedia users to see how this can be done. If nothing comes off it then we anyway have a plan for uploading these images. I maintain the SIFTS (pdbe.org/sifts) project which keeps up-to-date information on links between PDB and all other databases. This information is used by most databases (RCSB, Pfam, CATH, SCOP to name a few) so I suppose I am in a position to give all information about mappings and any changes to those mappings. So in summary, I agree with you that we can not use all the images and replace old ones but then will it be useful to create a new box with title "representative structure" and add the image. We could even make it (I don't know if that is possible) such that we add all the extra images that show mappings to other databases mentioned above. suggestions/thoughts? A2-25 (talk) 08:42, 26 February 2012 (UTC)
Citing patents
Thank you for your work on {{Citation/patent}} and {{cite patent}}. Note that I changed "dash" to "slash" in your post at Template talk:Citation/patent#Proposal to add DisplayedPublicationNumber parameter as I believe that is what you meant. Note also that patameters in {{cite patent}} are all lower case, so |DisplayedNumber= should probably be |displayednumber=. (Please respond either here or, better, at Template talk:Citation/patent.) HairyWombat 20:50, 11 March 2012 (UTC)
Later. I also corrected your link to discussion. HairyWombat 20:58, 11 March 2012 (UTC)
- I am rapdily loosing patience with your obsessive nitpicking. The purpose of the proposals (unindexed inventor parameter and DisplayedPublicationNumber) is to first to determine if there is support for these changes. If there is support, we can then make sure all the "i's are dotted and the t's are crossed". Please also keep in mind WP:TPO. Thank you. Boghog (talk) 07:30, 12 March 2012 (UTC)
WP:TPO states, "This page in a nutshell: Talk pages are for improving the encyclopedia, not for expressing personal opinions on a subject or an editor." I did not express a personal opinion on a subject or on an editor. What you call "nitpicking", I call accuracy; undotted 'i's and non-crossed 't's can cause confusion, and so are unhelpful when gathering support. I mentioned my corrections to your proposal here only as a courtesy. HairyWombat 17:10, 12 March 2012 (UTC)
Disambiguation link notification for March 17
Hi. When you recently edited SMC1A, you added a link pointing to the disambiguation page ATM (check to confirm | fix with Dab solver). Such links are almost always unintended, since a disambiguation page is merely a list of "Did you mean..." article titles. Read the FAQ • Join us at the DPL WikiProject.
It's OK to remove this message. Also, to stop receiving these messages, follow these opt-out instructions. Thanks, DPL bot (talk) 10:32, 17 March 2012 (UTC)
IUPHAR links
Have you managed to make any progress with the IUPHAR links?
regards Chidochangu (talk) 14:56, 28 March 2012 (UTC)
- Hi Chidochangu. The mapping of Wikipedia drug articles to IUPHAR pages is very difficult since Wikipedia does not have any database mechanism (although Wikidata may provide such functionality in the future).
- I was able to locate a number of Wikipedia drug articles that were not yet linked to IUPHAR. I have since then added these links. A mapping of Wikipedia drug articles to IUPHAR ligand IDs may be found here (please note that this list does not include mappings that were already in your list).
- I have also updated the {{IUPHAR}} template and activated the new links. A mapping of Wikipedia Gene Wiki articles (that link to the IUPHAR database) to Entrez Gene IDs may be found here. Boghog (talk) 15:51, 10 April 2012 (UTC)
- Hi Thank for your help with this list.. I will see what we can do to get the receprocal links set up on IUPHAR-db and will try to send an example to you soon... Chidochangu (talk) —Preceding undated comment added 08:39, 11 April 2012 (UTC).
- Great! Again, I apologize for taking so long but I have been busy in real life and determining the drugbox mappings was non-trivial. Thank you for your help in establishing the reciprocal links. Cheers. Boghog (talk) 18:53, 11 April 2012 (UTC)
WikiThanks

You are among the top 5% of most active Wikipedians this month! 66.87.2.119 (talk) 16:04, 31 March 2012 (UTC)
Image request

Hi Boghog! I'm preparing the article "GPCR oligomer" for the german wikipedia in my sandbox. I'd like to ask you whether you could make a sectional drawing of a model of a receptor heterodimer embedded in a membrane. --Hm20 (talk) 10:24, 1 April 2012 (UTC)
- Hi Hm20! I would be happy to help, but I am not exactly sure how to do this. If an experimental structure of a heterodimer were available, this would be very straight forward. Correct me if I am wrong, but I believe no such experimental structures exists. Lacking an experimental structure, do you known if some one has created a model of a heterodimeric structure? Lacking a heterodimer structure, are there any structures (experimental or modeled) of homodimers available (parallel, not anti-parallel)? Boghog (talk) 11:16, 1 April 2012 (UTC)
- Hi, The kappa opioid receptor PDB 4djh is a parallel homodimer in the crystal. It was published in Nature a couple of weeks ago (epub. don't think its in print yet PMID 22437504). Most GPCRs though tend to crystallise as head-to-tail dimers, which is biologically irrelevant. ~~A2-33
- Excellent! I take this one. Hm20 (talk) 17:11, 1 April 2012 (UTC)
- Wow, hot off the presses! Thanks A2-33! Hm20, I assume that you want something analogous to the image in TRPV. I will see what I can do. Boghog (talk) 18:08, 1 April 2012 (UTC)
- Initial attempt to the right. How does this look? Boghog (talk) 20:22, 1 April 2012 (UTC)
Thank you very much, Boghog! I love your drawings. Hm20 (talk) 20:56, 1 April 2012 (UTC)
- I just noticed that the 4djh crystal structure is of a artificial fusion protein between the κ-opioid receptor and lysozyme. In the original graphic, lysozyme appeared to be an intracellular domain which obviously is not right. I have updated the graphic to suppress the display of lysozyme. Sorry about that. Boghog (talk) 06:34, 7 April 2012 (UTC)
WikiProject Immunology
I see you have edited some of the pages within the scope of immunology. Please have a look at the proposal for a WikiProject Immunology WP:WikiProject Council/Proposals/Immunology and give your opinion (support or oppose). Thank you for your attention. Kinkreet~♥moshi moshi♥~ 09:28, 4 April 2012 (UTC)
Disambiguation link notification for April 5
Hi. When you recently edited GDF6, you added links pointing to the disambiguation pages Epidermis and Noggin (check to confirm | fix with Dab solver). Such links are almost always unintended, since a disambiguation page is merely a list of "Did you mean..." article titles. Read the FAQ • Join us at the DPL WikiProject.
It's OK to remove this message. Also, to stop receiving these messages, follow these opt-out instructions. Thanks, DPL bot (talk) 10:25, 5 April 2012 (UTC)
Resolving KCNMA1
Hi Boghog, I noticed your change to KCNMA1 and realized that there are partially redundant articles on this channel. I worked on this channel for 5 years during my PhD and participated the publication of it crystal structure 7 years later, so it's dear to my heart :) I'd like to propose merging BK channel and KCNMA1, and using 'KCNMA1' as the title. This will reduce redundancy and unify the Gene Wiki article nomenclature with KCNMB1, KCNMB2, etc. Do you have recommendations on how best to proceed? For example, after merging content, should I just add redirects to the old articles? AlexanderPico (talk) 17:49, 11 April 2012 (UTC)
- Hi Alexander. It is great to have some experts like yourself contributing to these Gene Wiki articles! Concerning Calcium-activated potassium channel subunit alpha-1, I have no objection renaming this article to KCNMA1. I am not so sure about merging the BK channel article into KCNMA1 however. The later is a gene/protein specific article while the former is about the alpha tetramer + optional beta subunits. I think it makes some sense to have one parent article that covers both alpha and beta subunits (BK channel) plus separate gene/protein specific articles (KCNMA1, KCNMB1, KCNMB2, KCNMB3, KCNMB4). One thing that probably should be moved is the {{Infobox protein family}} (Pfam calcium-activated BK potassium channel alpha subunit) from BK channel to KCNMA1. Thoughts? Boghog (talk) 18:37, 11 April 2012 (UTC)
- I had the same thought initially, but actually the entire BK article is only relevant to the alpha subunit and doesn't say anything substantial about the beta subunits. I was also persuaded by the fact that other articles and templates already provide overview of the complex and list the beta subunits: Calcium-activated_potassium_channel and Template:Ion_channels. AlexanderPico (talk) 19:22, 11 April 2012 (UTC)
- ^ PDB: 4DJH; Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC (2012). "Structure of the human κ-opioid receptor in complex with JDTic". Nature. doi:10.1038/nature10939. PMID 22437504.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link)
