Spindle apparatus

In cell biology, spindle apparatus refers to the subcellular structure that segregates chromosomes between daughter cells during cell division. It is also referred to as the mitotic spindle during mitosis or the meiotic spindle during meiosis.
While the spindle apparatus is composed of hundreds upon hundreds of proteins,[1] the fundamental machinery are the spindle microtubules. Attachment of microtubules to chromosomes is mediated by kinetochores, which actively monitor spindle formation and prevent premature anaphase onset. Microtubule polymerization and depolymerization dynamics drive chromosome congression. Depolymerization of microtubules generates tension at kinetochores;[2] bipolar attachment of sister kinetochores to microtubules emanating from opposite cell poles couples opposing tension forces, aligning chromosomes at the cell equator and poising them for segregation to daughter cells. Once every chromosome is bi-oriented, anaphase commences and cohesin, which couples sister chromatids, is severed, permitting the transit of the sister chromatids to opposite poles.
Spindle structure
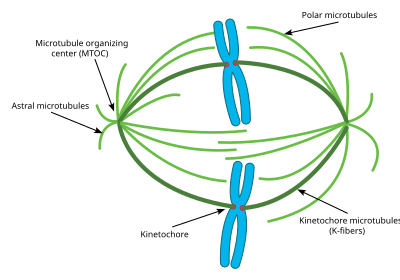
The cellular spindle apparatus includes the spindle microtubules, associated proteins, and any centrosomes or asters present at the spindle poles.[3] The spindle apparatus is vaguely ellipsoid in cross section and tapers at the ends. In the wide middle portion, known as the spindle midzone, antiparallel microtubules are bundled by kinesins. At the pointed ends, known as spindle poles, microtubules are nucleated by the centrosomes in most animal cells. Acentrosomal or anastral spindles lack centrosomes or asters at the spindle poles, respectively, and occur for example during gametogenesis in animals.[4] In fungi, spindles form between spindle pole bodies embedded in the nuclear envelope. Most plants lack centrosomes or spindle pole bodies and instead spindle microtubules are nucleated on their nuclear envelopes.[5]
Microtubule-associated proteins and spindle dynamics
The dynamic lengthening and shortening of spindle microtubules (Mitchison and Kirschner 1984) determines to a large extent the shape of the mitotic spindle and promotes the proper alignment of chromosomes at the spindle midzone. Microtubule-associated proteins (MAPs) associate with microtubules at the midzone and the spindle poles to regulate their dynamics. γ-tubulin is a specialized tubulin variant that assembles into a ring complex called γ-TuRC which nucleates polymerization of α/β tubulin heterodimers into microtubules. Recruitment of γ-TuRC to the pericentrosomal region stabilizes microtubule minus-ends and anchors them near the microtubule-organizing center.
The growing ends of microtubules are protected against catastrophe by the action of plus-end microtubule tracking proteins (+TIPs) to promote their association with kinetochores at the midzone. CLIP170 was shown to localize near microtubule plus-ends in HeLa cells [6] and to accumulate in kinetochores during prometaphase.[7] Although it remains unclear how CLIP170 recognizes plus-ends, it has been shown that its homologues protect against catastrophe and promote rescue,[8][9] suggesting a role for CLIP170 in stabilizing plus-ends and possibly mediating their direct attachment to kinetochores.[10] CLIP-associated proteins like CLASP1 in humans have also been shown to localize to plus-ends and the outer kinetochore as well as to modulate the dynamics of kinetochore microtubules (Maiato 2003). CLASP homologues in Drosophila, Xenopus, and yeast are required for proper spindle assembly; in mammals, CLASP1 and CLASP2 both contribute to proper spindle assembly and microtubule dynamics in anaphase.[11] Plus-end polymerization may be further moderated by EB1, which directly binds the growing ends of microtubules and coordinates the binding of other +TIPs.[12] Indeed, EB1 binds growing microtubules where they meet the kinetochore [13] and may not only promote plus-end stability but its growth. Its role during mitosis remains somewhat unclear, however, as its binding capacity is reduced in mitotic extracts.
Opposing the action of these microtubule-stabilizing proteins are a number of microtubule-depolymerizing factors which permit the dynamic remodeling of the mitotic spindle to promote chromosome congression and attainment of bipolarity. The kinesin-13 superfamily of MAPs contains a class of plus-end-directed motor proteins with associated microtubule depolymerization activity including the well-studied mammalian MCAK and Xenopus XKCM1. MCAK localizes to the growing tips of microtubules at kinetochores where it can trigger catastrophe in direct competition with stabilizing +TIP activity.[14] These proteins harness the energy of ATP hydrolysis to induce destabilizing conformational changes in protofilament structure that cause kinesin release and microtubule depolymerization.[15] Loss of their activity results in numerous mitotic defects.[14] Additional microtubule destabilizing proteins include Op18/stathmin and katanin which have roles in remodeling the mitotic spindle as well as promoting chromosome segregation during anaphase.[16]
The activities of these MAPs are carefully regulated to maintain proper microtubule dynamics during spindle assembly, with many of these proteins serving as Aurora and Polo-like kinase substrates.[16] As a testament to this, proper microtubule dynamics can be recapitulated in Xenopus egg extract by the balanced activity of the stabilizing factor XMAP215 and the destabilizing factor XKCM1.[17]
Organizing the spindle apparatus
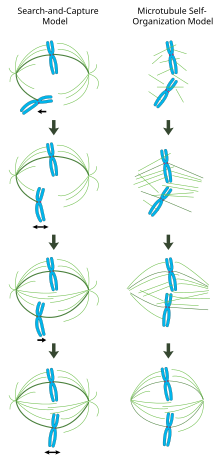
In a properly formed mitotic spindle, bi-oriented chromosomes are aligned along the equator of the cell with spindle microtubules oriented roughly perpendicular to the chromosomes, their plus-ends embedded in kinetochores and their minus-ends anchored at the cell poles. The precise orientation of this complex is required to ensure accurate chromosome segregation and to specify the cell division plane. However, it remains unclear how the spindle becomes organized. Two models predominate the field. In the search-and-capture model, the spindle is predominantly organized by the poleward separation of microtubule organizing centers (MTOCs). Spindle microtubules emanate from MTOCs and 'seek' out kinetochores; when they bind a kinetochore they become stabilized and exert tension on the chromosomes. In an alternative self assembly model, microtubules undergo acentrosomal nucleation among the condensed chromosomes. Constrained by cellular dimensions, lateral associations with antiparallel microtubules via motor proteins, and end-on attachments to kinetochores, microtubules naturally adopt a spindle-like structure with chromosomes aligned along the cell equator. Although these may be viewed as 'alternative' models, both phenomena likely contribute to the organization of the mitotic spindle.
Search-and-capture model
In this model, microtubules are nucleated at microtubule organizing centers and undergo rapid growth and catastrophe to 'search' the cytoplasm for kinetochores. Once they bind a kinetochore, they are stabilized and their dynamics are reduced. The newly mono-oriented chromosome oscillates in space near the pole to which it is attached until a microtubule from the opposite pole binds the sister kinetochore. This second attachment further stabilizes kinetochore attachment to the mitotic spindle. Gradually, the bi-oriented chromosome is pulled towards the center of the cell until microtubule tension is balanced on both sides of the centromere; the congressed chromosome then oscillates at the metaphase plate until anaphase onset releases cohesion of the sister chromatids.
In this model, microtubule organizing centers are localized to the cell poles, their separation driven by microtubule polymerization and 'sliding' of antiparallel spindle microtubules with respect to one another at the spindle midzone mediated by bipolar, plus-end-directed kinesins.[18][19] Such sliding forces may not only account for spindle pole separation early in mitosis, but also spindle elongation during late anaphase.
Self-organization of the mitotic spindle
In contrast to the search-and-capture mechanism in which MTOCs largely dictate the organization of the mitotic spindle, this model proposes that microtubules are nucleated acentrosomally near chromosomes and spontaneously assemble into anti-parallel bundles and adopt a spindle-like structure.[20] Classic experiments by Rebecca Heald show that functional mitotic spindles and nuclei form around DNA-coated beads incubated in Xenopus egg extracts and that bipolar arrays of microtubules are formed in the absence of kinetochores and MTOCs.[21] Indeed, it has also been shown that laser ablation of centrosomes in vertebrate cells inhibits neither spindle assembly nor chromosome segregation.[22] Under this scheme, the shape and size of the mitotic spindle are a function of the biophysical properties of the cross-linking motor proteins.[23]
Regulation of spindle assembly
Aurora A is required for proper spindle assembly and separation.[24] There have been identified many proteins necessary for the mitotic spindle assembly. Lamin B is not an essential protein for spindle assembly. It is only a component of the spindle matrix helping microtubule assembly, since proper mitotic spindle can be formed without it.[25]
Polo-like kinase, also known as PLK, especially PLK1 has important roles in the spindle maintenance by regulating microtubule dynamics.[26]
Mitotic spindle assembly checkpoint
The completion of spindle formation is a crucial transition point in the cell cycle called the spindle assembly checkpoint. If some chromosomes are not properly attached to the mitotic spindle by the time of this checkpoint, the onset of anaphase will be delayed.[27] Failure of this spindle assembly checkpoint can result in aneuploidy and may be involved in aging and the formation of cancer.[28] Abnormal mitotic spindles can produce tripolar mitosis. These are clearly abnormal cases and, if present, are considered definitive evidence that a tumor is malignant rather than benign. Such abnormalities are therefore often searched for in histological assays by pathologists when evaluating the potential malignancy of a tumor mass.
References
- ^ C. E. Walczak, R. Heald (2008). "Mechanisms of Mitotic Spindle Assembly and Function". International Review of Cytology. 265: 111–158.
- ^ E. Nogales, V. H. Ramey (1 November 2009). "Structure-function insights into the yeast Dam1 kinetochore complex". J of Cell Sci. 122: 3831–3836. doi:10.1242/jcs.004689.
{{cite journal}}: zero width space character in|doi=at position 9 (help) - ^ Campbell, Neil A. (2005). Biology, 7th Edition. San Francisco: Benjamin Cummings. pp. 221–224. ISBN 0-8053-7171-0.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Manandhar Gf, Schatten H, Sutovsky P (2005). "Centrosome reduction during gametogenesis and its significance". Biol. Reprod. 72 (1): 2–13. doi:10.1095/biolreprod.104.031245. PMID 15385423.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Schmit AC (2002). "Acentrosomal microtubule nucleation inzeff higherd plants". Int. Rev. Cytol. International Review of Cytology. 220: 257–89. doi:10.1016/S0074-7696(02)20008-X. ISBN 978-0-12-364624-8. PMID 12224551.
- ^ J.E. Rickard, T.E. Kreis (1990). "Identification of a novel nucleotide-sensitive microtubule-binding protein in HeLa cells". J Cell Biol. 110 (5): 1623–1633. doi:10.1083/jcb.110.5.1623. PMC 2200191. PMID 1970824.
- ^ D. Dujardin, U.I. Wacker, A. Moreau, T.A. Schroer, J.E. Rickard, J.R. DeMey (1998). "Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment". J Cell Biol. 141 (4): 849–862. doi:10.1083/jcb.141.4.849. PMC 2132766. PMID 9585405.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ D. Brunner, P. Nurse (2000). "CLIP-170-like tip1p spatially organizes microtubular dynamics in fission yeast". Cell. 102 (5): 695–704. doi:10.1016/S0092-8674(00)00091-X. PMID 11007487.
- ^ Y.A. Komarova, A.S. Kojima; et al. (2002). "Cytoplasmic linker proteins promote microtubule rescue in vivo". J Cell Biol. 159: 589–599. doi:10.1083/jcb.200208058. PMC 2173097. PMID 12446741.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ S. Goldstone, C. Reyes, G. Gay, T. Courthéoux, M. Dubarry; et al. (2010). "Tip1/CLIP-170 Protein Is Required for Correct Chromosome Poleward Movement in Fission Yeast". PLoS ONE. 5: e10634. doi:10.1371/journal.pone.0010634. PMC 2869355. PMID 20498706.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ A.L. Pereira, A.J. Pereira, A.R.R. Maia; et al. (1 October 2006). "Mammalian CLASP1 and CLASP2 Cooperate to Ensure Mitotic Fidelity by Regulating Spindle and Kinetochore Function". Mol Biol Cell. 17: 4526–4542. doi:10.1091/mbc.E06-07-0579. PMC 1635371. PMID 16914514.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ A. Akhmanova, M.O. Steinmetz (April 2008). "Tracking the ends: a dynamic protein network controls the fate of microtubule tips". Nat Rev Mol Cell Biol. 9 (4): 309–322. doi:10.1038/nrm2369. PMID 18322465.
- ^ J.S. Tirnauer, S. Grego, E.D. Salmon, T.J. Mitchison (1 October 2002). "EB1-microtubule interactions in Xenopus egg extracts: Role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules". Mol Biol Cell. 13 (10): 3614–3626. doi:10.1091/mbc.02-04-0210. PMC 129970. PMID 12388761.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b M.E. Tanenbaum, R.H. Medema, A. Akhmanova (2011). "Regulation of localization and activity of the microtubule depolymerase MCAK". Bioarchitecture. 1 (2): 80–87. doi:10.4161/bioa.1.2.15807. PMC 3158623. PMID 21866268.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ H. Niederstrasser, H. Salehi-Had, E.C. Gan, C. Walczak, E. Nogales (2002). "XKCM1 acts on a single protofilament and requires the C terminus of tubulin". J Mol Biol. 316 (3): 817–828. doi:10.1006/jmbi.2001.5360. PMID 11866534.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b H. Maiato, P Sampaio, C.E. Sunkel (2004). "Microtubule-associated proteins and their essential roles during mitosis". Int Rev Cytol. 241: 53–153. doi:10.1016/S0074-7696(04)41002-X. PMID 15548419.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ R. Tournebize, A. Popov, K. Kinoshita, A.J. Ashford; et al. (2000). "Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts". Nat Cell Biol. 2: 13–19. doi:10.1038/71330. PMID 10620801.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ J. McIntosh, S.C. Landis (1971). "The distribution of spindle microtubules during mitosis in cultured human cells". J Cell Biol. 49 (2): 468–497. doi:10.1083/jcb.49.2.468. PMC 2108320. PMID 19866774.
{{cite journal}}: Cite has empty unknown parameter:|1=(help) - ^ D.J. Sharp, K.L. McDonald, H.M. Brown; et al. (1999). "The bipolar kinesin, CLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles". J Cell Biol. 144 (1): 125–138. doi:10.1083/jcb.144.1.125. PMC 2148119. PMID 9885249.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ M.A. Hallen, S.A. Endow (2009). "Anastral spindle assembly: a mathematical model". Biophys J. 97 (8): 2191–2201. doi:10.1016/j.bpj.2009.08.008. PMC 2764103. PMID 19843451.
- ^ R. Heald, R. Tournebize; et al. (1996). "Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts". Nature. 382 (6590): 420–425. doi:10.1038/382420a0. PMID 8684481.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ A. Khodjakov, R.W. Cole, B.R. Oakley, C.L. Rieder (2000). "Centrosome-independent mitotic spindle formation in vertebrates". Curr Biol. 10 (2): 59–67. doi:10.1016/S0960-9822(99)00276-6. PMID 10662665.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ K.S. Burbank, T.J. Mitchison, D.S. Fisher (2007). "Slide-and-cluster models for spindle assembly". Curr Biol. 17 (16): 1373–1383. doi:10.1016/j.cub.2007.07.058. PMID 17702580.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ A.R. Barr, F. Gergely (2007). "Aurora A: The maker and breaker of spindle poles". J Cell Sci. 120: 2987–2996.
- ^ M. Y. Tsai, S. Wang, J. M. Heidinger, D. K. Shumaker, S. A. Adam, R. D. Goldman & Y. Zheng (31 March 2006). "A mitotic lamin B matrix induced by RanGTP required for spindle assembly". Science. 311 (5769): 1887–1193. doi:10.1126/science.1122771. PMID 16543417.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Peters, U., J. Cherian; et al. ([(2006)]). "Probing cell-division phenotype space and Polo-like kinase function using small molecules". Nat Chem Biol. 2 (11): 618–26. doi:10.1038/nchembio826. PMID 17028580.
{{cite journal}}: Check date values in:|date=(help); Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Raven, Peter H. (2005). Biology of Plants, 7th Edition. New York: W.H. Freeman and Company Publishers. p. 59. ISBN 0-7167-1007-2.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Baker DJ, Chen J, van Deursen JM (2005). "The mitotic checkpoint in cancer and aging: what have mice taught us?". Curr. Opin. Cell Biol. 17 (6): 583–9. doi:10.1016/j.ceb.2005.09.011. PMID 16226453.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
