Sobemovirus
| Sobemovirus | |
|---|---|
| |
| Genome of sesbania mosaic virus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Sobelivirales |
| Family: | Solemoviridae |
| Genus: | Sobemovirus |
Sobemovirus is a genus of viruses. Plants serve as natural hosts. There are currently 14 species in this genus including the type species Southern bean mosaic virus. Diseases associated with this genus include: mosaics and mottles.[1][2]
Structure
Viruses in Sobemovirus are non-enveloped, with icosahedral geometries, and T=3 symmetry. The diameter is around 30 nm.[1]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Sobemovirus | Icosahedral | T=3 | Non-enveloped | Linear | Monopartite |
Genome

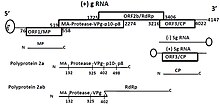
The genome is a single piece of linear, positive-sense, single-stranded RNA, 4100–5700 nucleotides in length.[3] The genome encodes five open reading frames: ORF1, ORFs 2a and 2b, ORF3 and ORFx.[4]
ORF1 encodes P1 which plays a role in suppression of silencing and virus movement.
ORFs 2a and 2b encode the replicational polyproteins P2a and P2ab. Translation of ORF2a from the genomic RNA is dependent on a leaky scanning mechanism.[citation needed]
ORF3 encodes the coat protein.[citation needed]
ORFx is conserved in all sobemoviruses. It overlaps the 5' end of ORF2a in the +2 reading frame and also extends some distance upstream of ORF2a. It lacks an AUG initiation codon and its expression is predicted to depend on low level initiation at near-cognate non-AUG codons, such as CUG, by a proportion of the ribosomes that are scanning the region between the ORF1 and ORF2a initiation codons. Its function is unknown but it appears to be essential for infection.[citation needed]
Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by penetration into the host cell. Replication follows the positive stranded RNA virus replication model. Positive stranded RNA virus transcription is the method of transcription. Translation takes place by leaky scanning, and -1 ribosomal frameshifting. The virus exits the host cell by tubule-guided viral movement. Plants serve as the natural host. The virus is transmitted via a vector (CFMV: insects). Transmission routes are vector and seed borne.[1]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Sobemovirus | Plants | None | Penetration | Viral Movement | Cytoplasm | Cytoplasm | Vector, Seed borne |
Taxonomy
Species included in the genus are:
- Blueberry shoestring virus
- Cocksfoot mottle virus
- Imperata yellow mottle virus
- Lucerne transient streak virus
- Rice yellow mottle virus
- Ryegrass mottle virus
- Sesbania mosaic virus
- Solanum nodiflorum mottle virus
- Southern bean mosaic virus
- Southern cowpea mosaic virus
- Sowbane mosaic virus
- Subterranean clover mottle virus
- Turnip rosette virus
- Velvet tobacco mottle virus
References
- ^ a b c "Viral Zone". ExPASy. Retrieved 15 June 2015.
- ^ ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- ^ Index of Viruses—Sobemovirus (2006). In: ICTVdB—The Universal Virus Database, version 4. Büchen-Osmond, C (Ed), Columbia University, New York, USA.[page needed]
- ^ Ling, Roger; Pate, Adrienne E; Carr, John P; Firth, Andrew E (2013). "An essential fifth coding ORF in the sobemoviruses". Virology. 446 (1–2): 397–408. doi:10.1016/j.virol.2013.05.033. PMC 3791421. PMID 23830075.
