Curacin A
 | |
| Identifiers | |
|---|---|
| |
| PubChem CID | |
| ChemSpider | |
| CompTox Dashboard (EPA) | |
| Chemical and physical data | |
| Formula | C23H35NOS |
| Molar mass | 373.60 g·mol−1 |
| 3D model (JSmol) | |
| |
| |
| (verify) | |
Curacin A is a hybrid polyketide synthase (PKS)/nonribosomal peptide synthase (NRPS) derived natural product produced isolated from the cyanobacterium Lyngbya majuscula.[1] Curacin A belongs to a family of natural products including jamaicamide, mupriocin, and pederin that have an unusual terminal alkene. Additionally, Curacin A contains a notable thiazoline ring and a unique cyclopropyl moiety, which is essential to the compound's biological activity.[1][2] Curacin A has been characterized as potent antiproliferative cytotoxic compound with notable anticancer activity for several cancer lines including renal, colon, and breast cancer.[2][3] Curacin A has been shown to interact with colchicine binding sites on tubulin, which inhibits microtubule polymerization, an essential process for cell division and proliferation.[1][4]
Biosynthesis
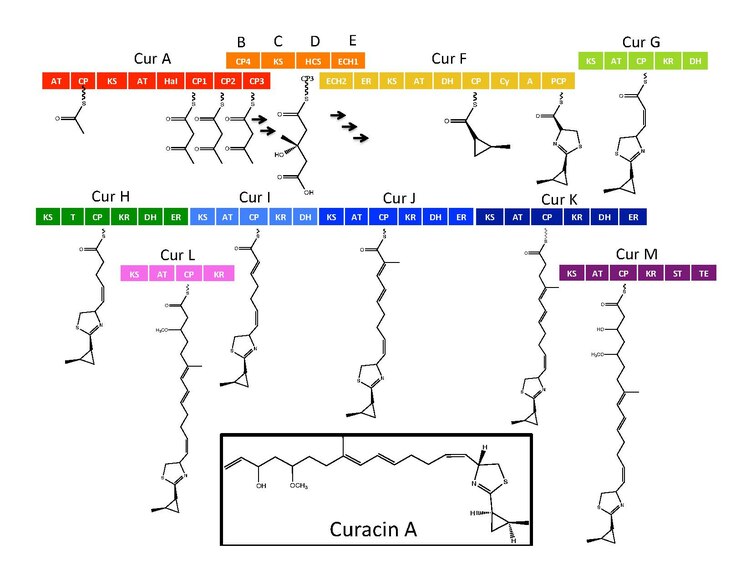
The synthetic enzymes for Curacin A are found in a gene cluster with 14 open reading frames (ORFs) with the nomenclature CurA through CurN.[1] Analysis of the pathway demonstrated the presence of one NRPS/PKS hybrid module located on CurF, one HMG-CoA synthase cassette located on CurD, and seven monomodular PKS modules.[1] CurA contains a unique GCN5-related N-acetyltransferase (GNAT) loading domain and an associated acyl carrier protein (ACP).[2] The loading module tethers an acetyl group to the ACP that then condenses with one of three tandem ACPs present in the adjacent module of CurA.[1][2][5] An hydroxymethylglutaryl-CoA synthase cassette (mevalonate pathway) catlyzes the formation of hydroxymethylglutaryl acid by the addition of an malonyl-CoA unit to the terminal ketide of the aceto-acetyl-ACP moiety of ACP1,ACP2, or ACP3.[5] subsequent enzymes, including a unique heme independent halogenase (HaI) catalyze the formation of a cyclopropyl ring.[1][5][6] A cysteine specific NRPS module located on CurF follows after cyclopropyl ring formation, and due to the activity of a cyclizing condensation domain, forms a thiazole ring attached to the cylcopropyl moiety from previous reactions in the pathway.[1][5][6] Seven standalone PKS modules follow to extend the growing polyketide chain with S-adenosyl methionine (SAM) dependent methylations occurring at positions 10 and 13.[1] A rare offloading strategy involving a sulfotransferase is employed by the final curacin synthase module. The sulfotransferase sulfates the hydroxyl group of carbon 15, which activates the molecule for decarboxylation and terminal alkene formation.[7]
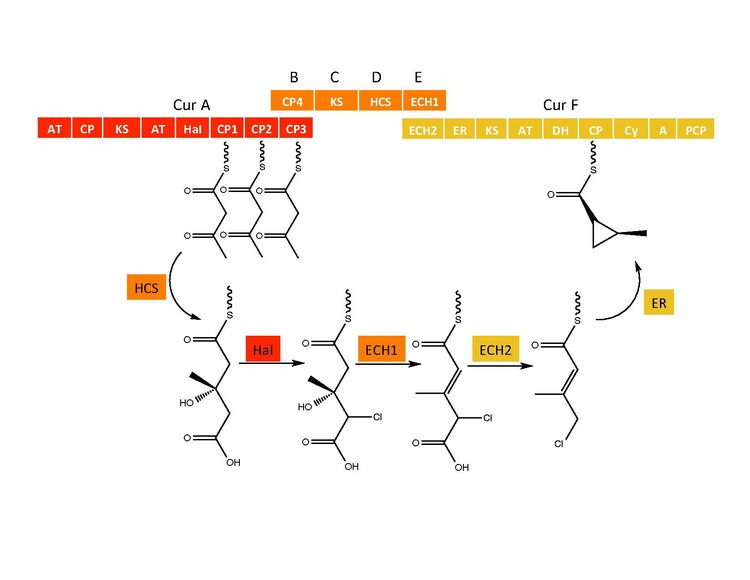
Cyclopropyl ring formation
The CurB (ACP), CurC (ketosynthase), and CurD (HMG-CoA reductase) are responsible for the formation of (S)HMG-ACP3.[6] HaI, from the CurA gene, is a unique non-heme halogenase that goes through a purported Fe(IV)=O intermediate to add a chlorine atom onto an unactivated carbon atom.[6] After chlorination, ECH1 acting as a dehydratates HMG-ACP3 to 3-methylgultaconyl-ACP3 and ECH2 performs the required decarboxylation.[6] Finally,an unusual ER catalyzed cyclization reaction, purported to go through a substitution like mechanism, forms the cyclopropane ring.[6] The added chlorine atom assists in the decarboxylation step and likely serves as the leaving group during cyclopropane ring formation.[6]
References
- ^ a b c d e f g h i Chang, Zunxue; Sitachitta, Namthip; Rossi, James V.; Roberts, Mary Ann; Flatt, Patricia M.; Jia, Junyong; Sherman, David H.; Gerwick, William H. (August 2004). "Biosynthetic Pathway and Gene Cluster Analysis of Curacin A, an Antitubulin Natural Product from the Tropical Marine Cyanobacterium". Journal of Natural Products. 67 (8): 1356–1367. doi:10.1021/np0499261.
- ^ a b c d Gu, Liangcai; Geders, Todd W.; Wang, Bo; Gerwick, William H.; Håkansson, Kristina; Smith, Janet L.; Sherman, David H. (2007). "GNAT-Like Strategy for Polyketide Chain Initiation". Science. 318: 970–974. doi:10.1126/science.1148790.
- ^ Verdier-Pinard, Pascal; Lai, Jing-Yu; Yoo, Hae-Dong; Yu, Jurong; Marquez, Brian; Nagle, Dale G.; Nambu, Mitch; White, James D.; Falck, J. R.; Gerwick, William H.; Day, Billy W.; Hamel, Ernest (1998). "Structure-Activity Analysis of the Interaction of Curacin A, the Potent Colchicine Site Antimitotic Agent, with Tubulin and Effects of Analogs on the Growth of MCF-7 Breast Cancer Cells". Molecular Pharmacology. 53 (1): 62–76. doi:10.1124/mol.53.1.62.
- ^ Blokhin, A. V.; Yoo, H. D.; Geralds, R. S.; Nagle, D. G.; Gerwick, W. H.; Hamel, E. (1995). "Characterization of the interaction of the marine cyanobacterial natural product curacin A with the colchicine site of tubulin and initial structure-activity studies with analogues". Molecular Pharmacology. 48: 523–531. PMID 7565634.
- ^ a b c d Gu, Liangcai; Eisman, Eli B.; Dutta, Somnath; Franzmann, Titus M.; Walter, Stefan; Gerwick, William H.; Skiniotis, Georgios; Sherman, David H. (2011). "Tandem Acyl Carrier Proteins in the Curacin Biosynthetic Pathway Promote Consecutive Multienzyme Reactions with a Synergistic Effect". Angewandte Chemie International Edition. 50 (12): 2795–2798. doi:10.1002/anie.201005280.
- ^ a b c d e f g Gu, Liangcai; Wang, Bo; Kulkarni, Amol; Geders, Todd W.; Grindberg, Rashel V.; Gerwick, Lena; Håkansson, Kristina; Wipf, Peter; Smith, Janet L.; Gerwick, William H.; Sherman, David H. (2009). "Metamorphic enzyme assembly in polyketide diversification". Nature. 459: 731–735. doi:10.1038/nature07870.
- ^ McCarthy, Jennifer Gehret; Eisman, Eli B.; Kulkarni, Sarang; Gerwick, Lena; Gerwick, William H.; Wipf, Peter; Sherman, David H.; Smith, Janet L. (2012). "Structural Basis of Functional Group Activation by Sulfotransferases in Complex Metabolic Pathways". ACS Chemical Biology. 7: 1994–2003. doi:10.1021/cb300385m.
{{cite journal}}: Cite has empty unknown parameter:|1=(help)