Dengue virus
| Dengue virus | |
|---|---|
| |
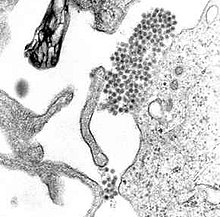
| A TEM micrograph showing dengue virus virions (the cluster of dark dots near the center). | |
| Virus classification | |
| Group: | Group IV ((+)ssRNA)
|
| Order: | Unassigned
|
| Family: | |
| Genus: | |
| Species: | Dengue virus
|
Dengue virus (DENV) is the cause of dengue fever. It is a mosquito-borne single positive-stranded RNA virus of the family Flaviviridae; genus Flavivirus.[1][2] Five serotypes of the virus have been found,[3] all of which can cause the full spectrum of disease.[1] Nevertheless, scientists are finding their understanding of dengue virus may be simplistic, as rather than distinct antigenic groups there appears to be a continuum.[4] This same study identified 47 strains of dengue virus.[5] Additionally, coinfection with and lack of rapid tests for zika virus and chikungunya complicate matters in real world infections.
Its genome is about 11000 bases of positive-sense single stranded RNA (ssRNA) that codes for three structural proteins (capsid protein C, membrane protein M, envelope protein E) and seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, NS5). It also includes short non-coding regions on both the 5' and 3' ends.[1][6]
Evolution
The dengue type 1 virus appears to have evolved in the early 19th century.[7] Based on the analysis of the envelope protein there are at least four genotypes (1 to 4). In 2013 a fifth serotype was reported.[3] The rate of nucleotide substitution for this virus has been estimated to be 6.5×10−4 per nucleotide per year, a rate similar to other RNA viruses. The American African genotype has been estimated to have evolved between 1907 and 1949. This period includes World Wars I and II, which were associated with considerable movement of populations and environmental disturbance, factors known to promote the evolution of new vector-borne viral species.
Life cycle
Until a few hundred years ago dengue virus was transmitted in sylvatic cycles in Africa and Asia between mosquitoes of the genus Aedes and non-human primates with rare emergences into human populations.[8][9] The global spread of dengue virus, however, has followed its emergence from sylvatic cycles and the primary life cycle now exclusively involves transmission between humans and Aedes mosquitoes.[10] Vertical transmission from mosquito to mosquito has also been observed in some vector species.[11]
Structures

E protein
The DENV E (envelope) protein, found as a dimer on the surface of the mature viral particle, is important in the initial attachment of this particle to the host cell. Each E protein monomer comprises three ectodomains, ED1 to ED3, and a trans-membrane segment. ED2 includes the dimerization interface, two glycosylation sites, and the peptide of fusion with the cellular membrane. ED3 is a continuous polypeptide segment; its fold is compact and immunoglobulin-like.[12][13] Dengue virus is transmitted by a mosquito known as Aedes. Several molecules which interact with the viral E protein (ICAM3-grabbing non-integrin,[14] CD209,[15] Rab 5,[16] GRP 78,[17] and the mannose receptor [18]) have been shown to be important factors mediating attachment and viral entry.[13] The membrane form of Ribosomal protein SA may also be involved in the attachment.[19] Recombinant domains of the E protein are used as well-defined antigens in the serological detection of antibodies directed against dengue virus and as immunogens in vaccine candidates.[20][21][22]
prM/M protein
The DENV prM (membrane) protein, which is important in the formation and maturation of the viral particle, consists of seven antiparallel β-strands stabilized by three disulfide bonds.[13]
The glycoprotein shell of the mature DENV virion consists of 180 copies each of the E protein and M protein. The immature virion starts out with the E and prM proteins forming 90 heterodimers that give a spiky exterior to the viral particle. This immature viral particle buds into the endoplasmic reticulum and eventually travels via the secretory pathway to the Golgi apparatus. As the virion passes through the trans-Golgi Network (TGN) it is exposed to low pH. This acidic environment causes a conformational change in the E protein which disassociates it from the prM protein and causes it to form E homodimers. These homodimers lie flat against the viral surface giving the maturing virion a smooth appearance. During this maturation pr peptide is cleaved from the M peptide by the host protease, furin. The M protein then acts as a transmembrane protein under the E-protein shell of the mature virion. The pr peptide stays associated with the E protein until the viral particle is released into the extracellular environment. This pr peptide acts like a cap, covering the hydrophobic fusion loop of the E protein until the viral particle has exited the cell.[13]
NS3 protein
The DENV NS3 is a serine protease, as well as an RNA helicase and RTPase/NTPase. The protease domain consists of six β-strands arranged into two β-barrels formed by residues 1–180 of the protein. The catalytic triad (His-51, Asp-75 and Ser-135), is found between these two β-barrels, and activity is dependent on the presence of the NS2B cofactor. This cofactor wraps around the NS3 protease domain and becomes part of the active site. The remaining NS3 residues (180–618), form the three subdomains of the DENV helicase. A six-stranded parallel β-sheet surrounded by four α-helices make up subdomains I and II, and subdomain III is composed of 4 α-helices surrounded by three shorter α-helices and two antiparallel β-strands.[13]
NS5 protein
The DENV NS5 protein is a 900 residue peptide with a methyltransferase domain at its N-terminal end (residues 1–296) and a RNA-dependent RNA polymerase (RdRp) at its C-terminal end (residues 320–900). The methyltransferase domain consists of an α/β/β sandwich flanked by N-and C-terminal subdomains. The DENV RdRp is similar to other RdRps containing palm, finger, and thumb subdomains and a GDD motif for incorporating nucleotides.[13]
Complexes between the E protein and neutralizing antibodies
Crystal structures of complexes between antibodies and either the ectodomain (sE) of the viral E protein or its domain 3 (ED3) have helped understand the molecular bases of the virus recognition and neutralization. Some of the epitopes are partially or totally innaccessible in the known structure of the mature virion. The corresponding antibodies are therefore assumed to bind to alternate or transitional conformations of the virus at 37 °C.
- The murine antibody E111 neutralizes DENV1. Its Fab and scFv fragments were crystallized in complex with the ED3 domain of DENV1. Its epitope is located around β-strands C and C' of ED3, and the intervening loop.[23]
- The murine antibody 1A1D-2 strongly neutralizes DENV1, DENV2 and DENV3. Its Fab fragment was crystallized in complex with the ED3 domain of DENV2. Its epitope straddles β-strands A and G of ED3.[24]
- The murine antibody 2H12 cross-reacts with all four DENV serotypes. It neutralizes the corresponding viruses, except DENV2. Its Fab fragment was crystallized in complex with the ED3 domains of DENV1, DENV3 and DENV4. Its epitope is located around the conserved AB loop of ED3.[25]
- The murine antibody 4E11 neutralizes all four DENV serotypes with varying efficacies. Its scFv fragment was crystallized in complex with the ED3 domains of the four DENV serotypes. Its epitope straddles β-strands A and G of ED3 as does the epitope of 1A1D-2.[26][27] The structures at 2.0 Å resolution have enabled one to analyze the roles of water molecules within the protein interfaces and the roles of somatic hypermutations outside of these interfaces in the interactions and cross-recognitions.[28]
- The chimpanzee antibody 5H2 potently neutralizes DENV4. Its Fab fragment was crystallized in complex with the sE protein of DENV4. Its epitope is included in domain 1 (ED1) of the E protein.[29]
- The human antibodies Ede1-C10, Ede2-A11 and Ede2-B7 potently neutralize all four DENV serotypes. Their Fab or scFv fragments were crystallized in complex with the sE protein of DENV2. The recognition determinants of these antibodies are at a serotype-invariant site in the E dimer interface and include the exposed side-chains of the E fusion loop and the two conserved glycan side-chains.[30]
Mechanism of Infection
- Dengue virus’ (DENV) E envelope protein binds to a cellular receptor. The exact nature of the cellular receptor has not been fully elucidated.
- DENV undergoes endocytosis. Acidification of the endosome leads to a conformational change of E, exposing a ‘fusion peptide’ sequence that facilitates fusion of the envelope with the endosomal membrane, releasing the virion capsid into the cytoplasm
- Uncoating occurs in the cytoplasm
- Host translational machinery (ribosomes) translates the (+)ssRNA into a single polypeptide
- Cellular and viral proteinases cleave the polypeptide into 10 proteins (E, M, C and 7 non-structural/enzymatic proteins) while embedded on the ER membrane
- As soon as functional RNA-dependent RNA polymerase is synthesised RNA replication can commence. Synthesis is asymmetrical, making 10 times more of the positive sense strand than the negative
- Assembly occurs on intracellular membranes which bud into the ER (forming the envelope from the ER membrane). Subsequent budding from the ER through the Golgi and into vesicles allows maturation via post-translational modifications eg glycosylation and pH transformational rearrangements
- Egress occurs via exocytosis[31]
Severe disease
The reason that some people suffer from more severe forms of dengue, such as dengue hemorrhagic fever, is multifactorial. Different strains of viruses interacting with people with different immune backgrounds lead to a complex interaction. Among the possible causes are cross-serotypic immune response, through a mechanism known as antibody-dependent enhancement, which happens when a person who has been previously infected with dengue gets infected for the second, third or fourth time. The previous antibodies to the old strain of dengue virus now interfere with the immune response to the current strain, leading paradoxically to more virus entry and uptake.[32]
Immune system interaction
In recent years, many studies have shown that flaviviruses, especially dengue virus has the ability to inhibit the innate immune response during the infection.[33][34] Indeed, the dengue virus has many nonstructural proteins that allow the inhibition of various mediators of the innate immune system response. These proteins act on two levels :
Inhibition of interferon signaling by blocking signal transducer
NS4B it is a small hydrophobic protein located in association with the endoplasmic reticulum. It may block the phosphorylation of STAT 1 after induction by interferons type I alpha, beta. In fact, the activity of Tyk2 kinase decreases with the dengue virus, so STAT 1 phosphorylation decreases too.[35] Therefore, the innate immune system response may be blocked. Thus there is no production of ISG. NS2A and NS4A cofactor may also take part in the STAT 1 inhibition.[36]
NS5 : the presence of this 105 kDa protein results in inactivation of STAT2 (via the signal transduction of the response to interferon) when it is expressed alone.[37] When NS5 is cleaved with NS4B by a protease (NS2B3) it can degrade STAT2. In fact, after the cleavage of NS5 by the protease, there is an E3 ligase association with STAT2, and the E3 ligase targets STAT2 for the degradation.[38][39]
Inhibition of the type I interferon response
NS2B3-b protease complex is a proteolytic core consisting of the last 40 amino acids of NS2B and the first 180 amino acids of NS3. Cleavage of the NS2B3 precursor activates the protease complex.[40]
This protease complex allows the inhibition of the production of type I interferon by reducing the activity of IFN-beta promoter: studies have shown that NS2B3 protease complex is involved in inhibiting the phosphorylation of IRF3.[41] A recent study shows that the NS2B3 protease complex inhibits (by cleaving) protein MITA which allows the IRF3 activation.[42]
Aedes aegypti D7 Saliva Protein
Dengue virus is transmitted by the mosquito species Aedes aegypti. Aedes aegypti produce saliva that contains over one hundred unique proteins, including the protein family D7.[43] Scientists use to believe that the Aedes aegypti saliva, when being transmitted, actually enhanced the dengue virus in the body. It was said that the mosquito’s spit could make the virus spread faster due to the weakened immune response of its host. However, a current study has found that the protein D7 hinders the virus transmission into the host cells.[43]
The immune responses of antibodies, that are trying to fight off the foreign virus, actually increase transmission and make the infection worse. Scientists found levels of protein D7 to be more prevalent in salivary glands of dengue-infected mosquitoes compared to those uninfected.[43] D7 is found in mosquito saliva and was thought to assist the process of blood feeding. Despite the prior assumptions, D7 can modulate the host cell and act against the virus to prevent virus infection.[43] Unfortunately D7 proteins provoke immune responses, which raise anti-D7 antibody levels. These antibodies inhibit the function of D7 proteins, which enhance transmission of dengue virus.
Vaccine research
Only one vaccine for dengue is currently approved in 3 countries (Brazil, Mexico, Philippines). Several vaccines are under development by private and public researchers.[44] Developing a vaccine against the disease is challenging. With four different serotypes of the dengue virus that can cause the disease, the vaccine must immunize against all four types to be effective.[3] Vaccination against only one serotype could possibly lead to severe dengue hemorrhagic shock (DHS) when infected with another serotype due to antibody-dependent enhancement. When infected with dengue virus, the immune system produces cross-reactive antibodies that provide immunity to that particular serotype. However, these antibodies are incapable of neutralizing other serotypes upon reinfection and actually increase viral replication. When macrophages consume the ‘neutralized’ virus, the virus is able replicate within the macrophage, causing disease. These cross-reactive, ineffective antibodies ease access of virus into macrophages, which induces more severe disease (dengue hemorrhagic fever, dengue shock syndrome). A common problem faced in dengue-endemic regions is when mothers become infected with dengue; after giving birth, offspring carry the immunity from their mother and are susceptible to hemorrhagic fever if infected with any of the other three serotypes.[45] One vaccine was in phase III trials in 2012 and planning for vaccine usage and effectiveness surveillance had started.[46]
In 2009 Sanofi-Pasteur started building a new facility in Neuville-sur-Saône (fr), a suburb of Lyon (France). This unit produces 4 serotypes vaccine for phase III trials. In September 2014 Sanofi-Pasteur CEO gave early results of the phase III trial efficacy study in Latin America. The efficacy per serotype (ST) varied widely, from 42.3% for ST2, 50.3% for ST1, 74.0% for ST3 and 77.7% for ST4. The full analysis of data from the phase III Latin American-Caribbean study will be reviewed by external experts before being published in a peer-reviewed scientific journal. Primary results has to be presented at the American Society of Tropical Medicine and Hygiene Annual Meeting, held November 2–6, 2014 in New Orleans.[47]
In September 2012, it was announced that one of the vaccines had not done well in clinical trials.[3]
References
- ^ a b c Rodenhuis-Zybert, Izabela A.; Wilschut, Jan; Smit, Jolanda M. (August 2010). "Dengue virus life cycle: viral and host factors modulating infectivity". Cellular and Molecular Life Sciences. 67 (16): 2773–2786. doi:10.1007/s00018-010-0357-z. ISSN 1420-682X. PMID 20372965.
- ^ WHO (2009). Dengue Guidelines for Diagnosis, Treatment, Prevention and Control (PDF). World Health Organization. ISBN 92-4-154787-1.
- ^ a b c d Normile, D (October 2013). "Tropical medicine. Surprising new dengue virus throws a spanner in disease control efforts". Science. 342 (6157): 415. Bibcode:2013Sci...342..415N. doi:10.1126/science.342.6157.415. PMID 24159024.
- ^ "A new understanding of dengue virus". ScienceDaily. September 18, 2015. Retrieved May 7, 2016.
- ^ "A Second Time Dengue Infection Can be Life-Threatening Says Research". NDTV. United Kingdom. September 18, 2015. Retrieved 2016-05-07.
- ^ Hanley, K.A.; Weaver, S.C., eds. (2010). Frontiers in Dengue Virus Research. Caister Academic. ISBN 978-1-904455-50-9.[page needed]
- ^ Patil JA, Cherian S, Walimbe AM, et al. (August 2011). "Evolutionary dynamics of the American African genotype of dengue type 1 virus in India (1962–2005)". Infection, Genetics and Evolution. 11 (6): 1443–8. doi:10.1016/j.meegid.2011.05.011. PMID 21632029.
- ^ "Dengue virus". Pathogen Information (PathInfo). Virginia Bioinformatics Institute, Virginia Tech.
- ^ Holmes, EC; Twiddy, SS (May 2003). "The origin, emergence and evolutionary genetics of dengue virus". Infection, Genetics and Evolution. 3 (1): 19–28. doi:10.1016/s1567-1348(03)00004-2. PMID 12797969.
- ^ Halstead, SB (1988). "Pathogenesis of dengue: challenges to molecular biology". Science. 239 (4839): 476–481. Bibcode:1988Sci...239..476H. doi:10.1126/science.3277268. PMID 3277268. Retrieved 23 February 2013.
- ^ Haddow, AD; Guzman, H; Popov, VL; Wood, TG; Widen, SG; Haddow, AD; Tesh, RB; Weaver, SC (Jun 5, 2013). "First isolation of Aedes flavivirus in the Western Hemisphere and evidence of vertical transmission in the mosquito Aedes (Stegomyia) albopictus (Diptera: Culicidae)". Virology. 440 (2): 134–9. doi:10.1016/j.virol.2012.12.008. PMID 23582303.
- ^ Modis, Y; Ogata, S; Clements, D; Harrison, SC (Jun 2003). "A ligand-binding pocket in the dengue virus envelope glycoprotein". Proc Natl Acad Sci USA. 100 (12): 6986–6991. doi:10.1073/pnas.0832193100. PMID 12759475.
- ^ a b c d e f Perera R, Kuhn RJ (August 2008). "Structural proteomics of dengue virus". Current Opinion in Microbiology. 11 (4): 369–77. doi:10.1016/j.mib.2008.06.004. PMC 2581888. PMID 18644250.
- ^ Tassaneetrithep B, Burgess TH, Granelli-Piperno A, et al. (April 2003). "DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells". J. Exp. Med. 197 (7): 823–9. doi:10.1084/jem.20021840. PMC 2193896. PMID 12682107.
- ^ Krishnan MN, Sukumaran B, Pal U, et al. (May 2007). "Rab 5 is required for the cellular entry of dengue and West Nile viruses". J. Virol. 81 (9): 4881–5. doi:10.1128/JVI.02210-06. PMC 1900185. PMID 17301152.
- ^ Jindadamrongwech, S.; Thepparit, C.; Smith, D. R. (May 2004). "Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2". Archives of Virology. 149 (5): 915–927. doi:10.1007/s00705-003-0263-x. ISSN 0304-8608. PMID 15098107.
- ^ Miller JL, de Wet BJ, deWet BJ, et al. (February 2008). "The mannose receptor mediates dengue virus infection of macrophages". PLoS Pathog. 4 (2): e17. doi:10.1371/journal.ppat.0040017. PMC 2233670. PMID 18266465.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Zidane, N; Ould-Abeih, MB; Petit-Topin, I; Bedouelle, H (Dec 2012). "The folded and disordered domains of human ribosomal protein SA have both idiosyncratic and shared functions as membrane receptors". Biosci Rep. 33 (1): 113–124. doi:10.1042/BSR20120103. PMID 23137297.
- ^ Zidane, N; Dussart, P; Bremand, L; Bedouelle, H (Jul 2013). "Cross-reactivities between human IgMs and the four serotypes of dengue virus as probed with artificial homodimers of domain-III from the envelope proteins". BMC Infect Dis. 13: 302. doi:10.1186/1471-2334-13-302. PMID 23815496.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Zidane, N; Dussart, P; Bremand, L; Villani, ME; Bedouelle, H (Jun 2013). "Thermodynamic stability of domain III from the envelope protein of flaviviruses and its improvement by molecular design". Protein Eng Des Sel. 26 (6): 389–399. doi:10.1093/protein/gzt010. PMID 23479674.
- ^ Brandler, S; Ruffie, C; Najburg, V; Frenkiel, MP; Bedouelle, H; Desprès, P; Tangy, F (Sep 2010). "Pediatric measles vaccine expressing a dengue tetravalent antigen elicits neutralizing antibodies against all four dengue viruses". Vaccine. 28 (41): 6730–6739. doi:10.1016/j.vaccine.2010.07.073. PMID 20688034.
- ^ Austin, SK; Dowd, KA; Shrestha, B; Nelson, CA; Edeling, MA; Johnson, S; Pierson, TC; Diamond, MS; Fremont, DH (2012). "Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope". PLoS Pathog. 8 (10): e1002930. doi:10.1371/journal.ppat.1002930. PMID 23055922.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lok, SM; Kostyuchenko, V; Nybakken, GE; Holdaway, HA; Battisti, AJ; Sukupolvi-Petty, S; Sedlak, D; Fremont, DH; Chipman, PR; Roehrig, JT; Diamond, MS; Kuhn, RJ; Rossmann, MG (Mar 2008). "Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins". Nat Struct Mol Biol. 15 (3): 312–317. doi:10.1038/nsmb.1382. PMID 18264114.
- ^ Midgley, CM; Flanagan, A; Tran, HB; Dejnirattisai, W; Chawansuntati, K; Jumnainsong, A; Wongwiwat, W; Duangchinda, T; Mongkolsapaya, J; Grimes, JM; Screaton, GR (May 2012). "Structural analysis of a dengue cross-reactive antibody complexed with envelope domain III reveals the molecular basis of cross-reactivity". J Immunol. 188 (10): 4971–4979. doi:10.4049/jimmunol.1200227. PMID 22491255.
- ^ Lisova, O; Hardy, F; Petit, V; Bedouelle, H (Sep 2007). "Mapping to completeness and transplantation of a group-specific, discontinuous, neutralizing epitope in the envelope protein of dengue virus". J Gen Virol. 88 (9): 2387–2397. doi:10.1099/vir.0.83028-0. PMID 17698647.
- ^ Cockburn, JJ; Navarro Sanchez, ME; Fretes, N; Urvoas, A; Staropoli, I; Kikuti, CM; Coffey, LL; Arenzana Seisdedos, F; Bedouelle, H; Rey, FA (Feb 2012). "Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody". Structure. 20 (2): 303–314. doi:10.1016/j.str.2012.01.001. PMID 22285214.
- ^ Lisova, O; Belkadi, L; Bedouelle, Hugues (Apr 2014). "Direct and indirect interactions in the recognition between a cross-neutralizing antibody and the four serotypes of dengue virus". J Mol Recognit. 27 (4): 205–214. doi:10.1002/jmr.2352. PMID 24591178.
- ^ Cockburn, JJ; Navarro Sanchez, ME; Goncalvez, AP; Zaitseva, E; Stura, EA; Kikuti, CM; Duquerroy, S; Dussart, P; Chernomordik, LK; Lai, CJ; Rey, FA (Feb 2012). "Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus". EMBO J. 31 (3): 767–779. doi:10.1038/emboj.2011.439. PMID 22139356.
- ^ Rouvinski, A; Guardado-Calvo, P; Barba-Spaeth, G; Duquerroy, S; Vaney, MC; Kikuti, CM; Navarro Sanchez, ME; Dejnirattisai, W; Wongwiwat, W; Haouz, A; Girard-Blanc, C; Petres, S; Shepard, WE; Desprès, P; Arenzana-Seisdedos, F; Dussart, P; Mongkolsapaya, J; Screaton, GR; Rey, FA (Apr 2015). "Recognition determinants of broadly neutralizing human antibodies against dengue viruses". Nature. 520 (7545): 109–113. doi:10.1038/nature14130. PMID 25581790.
- ^ Acheson, Nicholas H. (2011). Fundamentals of Molecular Virology, 2nd ed. Wiley.
- ^ Dejnirattisai W, Jumnainsong A, Onsirisakul N, et al. (May 2010). "Cross-reacting antibodies enhance dengue virus infection in humans". Science. 328 (5979): 745–8. Bibcode:2010Sci...328..745D. doi:10.1126/science.1185181. PMID 20448183.
- ^ Diamond MS (September 2009). "Mechanisms of evasion of the type I interferon antiviral response by flaviviruses". J. Interferon Cytokine Res. 29 (9): 521–30. doi:10.1089/jir.2009.0069. PMID 19694536.
- ^ Jones M, Davidson A, Hibbert L, et al. (May 2005). "Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression". J. Virol. 79 (9): 5414–20. doi:10.1128/JVI.79.9.5414-5420.2005. PMC 1082737. PMID 15827155.
- ^ Ho LJ, Hung LF, Weng CY, et al. (June 2005). "Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell". Journal of Immunology. 174 (12): 8163–72. doi:10.4049/jimmunol.174.12.8163. PMID 15944325.
- ^ Munoz-Jordan, J. L.; Sanchez-Burgos, G. G.; Laurent-Rolle, M.; Garcia-Sastre, A. (November 2003). "Inhibition of interferon signaling by dengue virus". Proceedings of the National Academy of Sciences. 100 (24): 14333–14338. Bibcode:2003PNAS..10014333M. doi:10.1073/pnas.2335168100. ISSN 0027-8424. PMC 283592. PMID 14612562.
- ^ Ashour, J.; Laurent-Rolle, M.; Shi, P.-Y.; Garcia-Sastre, A. (June 2009). "NS5 of Dengue Virus Mediates STAT2 Binding and Degradation". Journal of Virology. 83 (11): 5408–5418. doi:10.1128/JVI.02188-08. ISSN 0022-538X. PMC 2681973. PMID 19279106.
- ^ Mazzon, Michela; Jones, Meleri; Davidson, Andrew; Chain, Benjamin; Jacobs, Michael (October 2009). "Dengue Virus NS5 Inhibits Interferon‐α Signaling by Blocking Signal Transducer and Activator of Transcription 2 Phosphorylation". The Journal of Infectious Diseases. 200 (8): 1261–1270. doi:10.1086/605847. ISSN 0022-1899. PMID 19754307.
- ^ Morrison, Juliet; Aguirre, Sebastian; Fernandez-Sesma, Ana (March 2012). "Innate Immunity Evasion by Dengue Virus". Viruses. 4 (12): 397–413. doi:10.3390/v4030397. ISSN 1999-4915. PMC 3347034. PMID 22590678.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Yusof, Rohana; Clum, Stephen; Wetzel, Mary; Murthy, H. M. Krishna; Padmanabhan., R. (April 2000). "Purified NS2B/NS3 Serine Protease of Dengue Virus Type 2 Exhibits Cofactor NS2B Dependence for Cleavage of Substrates with Dibasic Amino Acids in Vitro". Journal of Biological Chemistry. 275 (14): 9963–9969. doi:10.1074/jbc.275.14.9963. ISSN 0021-9258. PMID 10744671.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Rodriguez-Madoz, J. R.; Belicha-Villanueva, A.; Bernal-Rubio, D.; Ashour, J.; Ayllon, J.; Fernandez-Sesma, A. (October 2010). "Inhibition of the Type I Interferon Response in Human Dendritic Cells by Dengue Virus Infection Requires a Catalytically Active NS2B3 Complex". Journal of Virology. 84 (19): 9760–9774. doi:10.1128/JVI.01051-10. ISSN 0022-538X. PMC 2937777. PMID 20660196.
- ^ Yu, Chia-Yi; Chang, Tsung-Hsien; Liang, Jian-Jong; Chiang, Ruei-Lin; Lee, Yi-Ling; Liao, Ching-Len; Lin, Yi-Ling (June 2012). Diamond, Michael S. (ed.). "Dengue Virus Targets the Adaptor Protein MITA to Subvert Host Innate Immunity". PLoS Pathogens. 8 (6): e1002780. doi:10.1371/journal.ppat.1002780. ISSN 1553-7374. PMC 3386177. PMID 22761576.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d Conway, Michael; Londono-Renteria, Berlin (September 15, 2016). "Aedes aegypti D7 Saliva Protein Inhibits Dengue Virus Infection". Neglected Tropical Diseases. Public Library of Sciences. doi:10.1371/journal.pntd.0004941.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Vaccine Development, Dengue Vaccine Initiative, November 2012, accessed November 5, 2013
- ^ Schmaljohn, Alan L.; McClain, David (1996). "Ch. 54: Alphaviruses (Togaviridae) and Flaviviruses (Flaviviridae)". In Baron S (ed.). Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 0-9631172-1-1. NBK7633.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ Torresi J, Tapia-Conyer R, Margolis H (2013). "Preparing for dengue vaccine introduction: recommendations from the 1st dengue v2V International Meeting". PLoS Negl Trop Dis. 7 (9): e2261. doi:10.1371/journal.pntd.0002261. PMC 3784468. PMID 24086776.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Sanofi Pasteur Dengue Vaccine Aces Second Phase III Trial". GEN News Highlights. Genetic Engineering & Biotechnology News. 3 September 2014.
External links
- 3D electron microscopy structures of dengue virus from the EM Data Bank(EMDB)
- "Brazil releases 'good' mosquitoes to fight dengue fever". BBC News Latin America & Caribbean. 24 September 2014.
- "Dengue virus". NCBI Taxonomy Browser. 12637.
