User:Cherrysu/sandbox
 | This is a user sandbox of Cherrysu. You can use it for testing or practicing edits. This is not the sandbox where you should draft your assigned article for a dashboard.wikiedu.org course. To find the right sandbox for your assignment, visit your Dashboard course page and follow the Sandbox Draft link for your assigned article in the My Articles section. |
| PreQ1 riboswitch | |
|---|---|
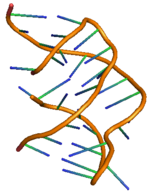
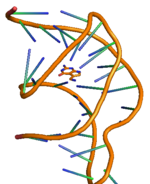
 Predicted secondary structure and sequence conservation of PreQ1 | |
| Identifiers | |
| Symbol | PreQ1 |
| Rfam | RF00522 |
| Other data | |
| RNA type | Cis-reg; riboswitch |
| Domain(s) | Bacteria |
| SO | SO:0000035 |
| PDB structures | PDBe 2L1V |
The PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine (Q) from GTP.[1] PreQ1 (pre-queuosine1) is an intermediate in the queuosine pathway, and preQ1 riboswitch, as a type of riboswitch, is an RNA element that binds preQ1. The preQ1 riboswitch is distinguished by its unusually small aptamer, compared to other riboswitches. Its atomic-resolution three-dimensional structure has been determined, with the PDB ID 2L1V.[2][3]
PreQ1 Classification
[edit]Three subcategories of the PreQ1 riboswitch exist: preQ1-I, preQ1-II, and preQ1-III. PreQ1-I has a distinctly small aptamer, ranging from 25 to 45 nucleotides long, [4]compared to the structures of PreQ1-II riboswitch and preQ1-III riboswitch. PreQ1-II riboswitch, only found in Lactobacillales, has a larger and more complex consensus sequence and structure than preQ1-I riboswitch, with an average of 58 nucleotides composing its aptamer, which forms as many as five base-paired substructures.[5] PreQ1-III riboswitch has a distinct structure and is also larger in aptamer size than preQ1-I riboswitch, ranging from sizes ranging from 33 to 58 nucleotides. PreQ1-III riboswitch has an atypically organized pseudoknot that does not appear to incorporate its downstream expression platform at its ribosome binding site (RBS).[6]
History
[edit]While preQ1 was first discovered as an anticodon sequence of tRNAs from E.coli in 1972[7], preQ1 riboswitch was not first found until 2004[8] and recognized even later.[9] The first reported preQ1 riboswitch was located in the leader of the Bacillus subtilis ykvJKLM (queCDEF) operon which encodes four genes necessary for queuosine production[8]. In this organism, PreQ1 binding to the riboswitch aptamer is thought to induce premature transcription termination within the leader to down-regulate expression of these genes. Later on, preQ1 riboswitch was identified as a conserved sequence on the 5' UTR of genes in many gram-positive bacteria and was proved to be associated with synthesis of preQ1[9].
In 2008, a second class of preQ1 riboswitch (PreQ1-II riboswitches) was also found as a representative of the COG4708 RNA motif from Streptococcus pneumoniae R6.[10] Although PreQ1-II riboswitch also works as queuosine biosynthetic intermediate, the structural and molecular recognition characteristics are distinct from preQ1-I riboswitch, indicating that natural aptamers utilizing different structures to bind the same metabolite may be more common than is currently known.[10]
Structure and Function
[edit]

PreQ1 riboswitch has two stems and three loops, and its detailed structure has been shown on the right.[11] The riboswitching action of preQ1 riboswitches in bacteria is regulated by binding of metabolite preQ1 to the aptamer region leading to structural changes in the messenger RNA (mRNA) that governs the downstream genetic regulation.[12] The preQ1 riboswitch structure adopts a compact H-type pseudoknot, which makes it quite different from other purine based riboswitches.[12] The preQ1 ligand is buried in the pseudoknot core and stabilized through intercalation between helical stacks and hydrogen bond interaction with heteroatoms. In absence of preQ1, the P2 tail region is away from the P2 loop region and hence the riboswitch is observed to be in undocked (partially docked) state, whereas on the binding of preQ1 to the riboswitch results the two P2 regions to come closer causing a complete docking of riboswitch. This docking and undocking mechanism of riboswitch with the change in concentration of ligand preQ1 is observed to control the signaling of gene regulation, commonly known as “ON” or “OFF” signaling for gene expression.[11][13] The docking and undocking mechanism are observed to be affected not only by the ligand, but also with other factors like Mg salt.[14] Like any other riboswitch, the two most common types of gene regulation mediated by preQ1 riboswitch are through transcription attenuation or inhibition of translation initiation. Ligand binding to the transcriptional riboswitch in bacterial causes modification in the structure of riboswitch unit, which leads to hindrance in the activity of RNA polymerase causing attenuation of transcription. Similarly, binding of the ligand to the translational riboswitch causes modification in the secondary structure of riboswitch unit leading to hindrance for ribosome binding and hence inhibiting translational initiation.
Transcriptional regulation
[edit]PreQ1 mediated transcriptional attenuation is controlled by the dynamic switching of anti-terminator and terminator hairpin in the riboswitch.[11] For preQ1 riboswitch from bacteria Bacillus subtilis (Bsu), the anti-terminator is predicted to be less stable than the terminator, as the addition of preQ1 shifts the equilibrium significantly towards the formation of terminator.[11] In presence of preQ1, the 3’ end of the adenine rich tail domain pairs with the center of P1 hairpin loop to form an H-type pseudoknot[11]. In the native mRNA structure, binding of preQ1 to the aptamer region in the riboswitch leads to the formation of a terminator hairpin which causes RNA polymerase to stop transcription, a process which is commonly known as OFF- regulation of genetic expression or transcription termination.[13]
Translation regulation
[edit]Translation of protein in prokaryotes is initiated by binding of 30S ribosomal subunit to the Shine-Dalgarno (SD) sequence in mRNA. PreQ1 mediated inhibition of translational regulation is controlled by blocking the Shine-Dalgarno sequence of mRNA to prevent binding of ribosome to mRNA for translation. Binding of preQ1 to the aptamer domain promotes the sequestration of a part of SD sequence at the 5’ end to the P2 stem of the aptamer domain causing inaccessibility of the SD sequence[11][15]. The translational riboswitch from bacteria Thermoanaerobacter tengcongensis (Tte) is observed to be transiently closed (pre-docked) in absence of preQ1, whereas in presence of preQ1 a fully docked state is adopted. This docking/undocking equilibrium is not only regulated by the concentration of ligand but also by the concentration of Mg salt.[14][16] The unavailability of SD sequence due to the formation of pseudoknot in presence of preQ1 shows the OFF-regulation of genetic expression in translational riboswitch or inhibition of translational initiation.
Physiological relevance in bacterial gene regulation
[edit]PreQ1 riboswitch activity in Tte bacteria can be measured by the levels of two proteins that are in the coding region of the Tte mRNA, which are TTE1564 and TTE1563[17]. Proteins downstream of the preQ1 riboswitch biosynthesize a nucleobase called queuine and a nucleoside queuosine are inhibited by the activation of the preQ1 riboswitch. Queuine is involved in the anticodon sequence of certain tRNA[18]. In bacteria, the hyper-modified nucleobase queuine takes up the first anticodon position, or its wobble position in the tRNA of Asparagine, Aspartic acid, Histidine, and Tyrosine[19]. In bacteria the enzyme tRNA-guanine transglycosylase (TGT) catalyzes the swap of a guanine in position 34 of the tRNA with queuine into the first anitcodon position[16][17]. Eukaryae incorporate queuine into RNA, whereas eubacteria incorporate preQ1, which then undergoes modification to yield queuine[18].Since queuosine is exclusively produced in bacteria, eukaryotic organisms must obtain their supply of queuosine or its nucleobase queuine from their diet or bacteria from their gut microflora. The implication of a deficiency of queuine or queuosine is an inability to make queuosine-modified tRNA, and furthermore, the inability of the cell to convert phenylalanine to tyrosine[20].
See Also
[edit]References
[edit]- ^ Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, Iwata-Reuyl D, Breaker RR (2007). "A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain". Nat Struct Mol Biol. 14 (4): 308–317. doi:10.1038/nsmb1224. PMID 17384645.
- ^ Klein DJ, Edwards TE, Ferré-D'Amaré AR (March 2009). "Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase". Nat. Struct. Mol. Biol. 16 (3): 343–344. doi:10.1038/nsmb.1563. PMC 2657927. PMID 19234468.
- ^ Kang M, Peterson R, Feigon J (March 2009). "Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA". Mol. Cell. 33 (6): 784–90. doi:10.1016/j.molcel.2009.02.019. PMID 19285444.
- ^ "RIBOSWITCHES: CLASSIFICATION, FUNCTION and INSILICO APPROACH". International Journal of Pharma Sciences and Research (IJPSR). 1(9), 2010, 414.
- ^ "Structural, Functional, and Taxonomic Diversity of Three PreQ1 Riboswitch Classes". Chemistry & Biology. 21, Issue 7, 17 July 2014, Pages 880–889.
- ^ Liberman, Joseph A.; Suddala, Krishna C.; Aytenfisu, Asaminew; Chan, Dalen; Belashov, Ivan A.; Salim, Mohammad; Mathews, David H.; Spitale, Robert C.; Walter, Nils G. (2015-07-07). "Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics". Proceedings of the National Academy of Sciences. 112 (27): E3485–E3494. doi:10.1073/pnas.1503955112. ISSN 0027-8424. PMC 4500280. PMID 26106162.
- ^ Harada, Fumio; Nishimura, Susumu (January 1972). "Possible anticodon sequences of tRNAHis, tRNAAsn, and tRNAAsp from Escherichia coli. Universal presence of nucleoside O in the first position of the anticodons of these transfer ribonucleic acid". Biochemistry. 11 (2): 301–308. doi:10.1021/bi00752a024.
- ^ a b Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V (2004). "Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine". J. Biol. Chem. 279 (8): 6280–6285. doi:10.1074/jbc.M310858200. PMID 14660578.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Roth, Adam; Winkler, Wade C.; Regulski, Elizabeth E.; Lee, Bobby W. K.; Lim, Jinsoo; Jona, Inbal; Barrick, Jeffrey E.; Ritwik, Ankita; Kim, Jane N. (2007-04-01). "A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain". Nature Structural & Molecular Biology. 14 (4): 308–317. doi:10.1038/nsmb1224. ISSN 1545-9993. PMID 17384645.
- ^ a b Meyer, Michelle M.; Roth, Adam; Chervin, Stephanie M.; Garcia, George A.; Breaker, Ronald R. (2008-04-01). "Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria". RNA. 14 (4): 685–695. doi:10.1261/rna.937308. ISSN 1355-8382.
- ^ a b c d e f Eichhorn, Catherine D.; Kang, Mijeong; Feigon, Juli (2014-10-01). "Structure and function of preQ1 riboswitches". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. Riboswitches. 1839 (10): 939–950. doi:10.1016/j.bbagrm.2014.04.019. PMC 4177978. PMID 24798077.
- ^ a b Serganov, Alexander; Patel, Dinshaw J. (2012-05-11). "Metabolite Recognition Principles and Molecular Mechanisms Underlying Riboswitch Function". Annual Review of Biophysics. 41 (1): 343–370. doi:10.1146/annurev-biophys-101211-113224. ISSN 1936-122X. PMC 4696762. PMID 22577823.
- ^ a b Rieder, Ulrike; Kreutz, Christoph; Micura, Ronald (2010-06-15). "Folding of a transcriptionally acting PreQ1 riboswitch". Proceedings of the National Academy of Sciences. 107 (24): 10804–10809. doi:10.1073/pnas.0914925107. ISSN 0027-8424.
- ^ a b Suddala, Krishna C.; Rinaldi, Arlie J.; Feng, Jun; Mustoe, Anthony M.; Eichhorn, Catherine D.; Liberman, Joseph A.; Wedekind, Joseph E.; Al-Hashimi, Hashim M.; Brooks, Charles L. (2013-12-01). "Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure". Nucleic Acids Research. 41 (22): 10462–10475. doi:10.1093/nar/gkt798. ISSN 0305-1048. PMC 3905878. PMID 24003028.
- ^ Eichhorn, Catherine D.; Kang, Mijeong; Feigon, Juli (2014-10-01). "Structure and function of preQ1 riboswitches". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. Riboswitches. 1839 (10): 939–950. doi:10.1016/j.bbagrm.2014.04.019. PMC 4177978. PMID 24798077.
- ^ a b Suddala, Krishna C.; Wang, Jiarui; Hou, Qian; Walter, Nils G. (2015-11-11). "Mg2+ Shifts Ligand-Mediated Folding of a Riboswitch from Induced-Fit to Conformational Selection". Journal of the American Chemical Society. 137 (44): 14075–14083. doi:10.1021/jacs.5b09740. ISSN 0002-7863.
- ^ a b E., Lund, Paul (2015-01-01). "Interactions between the Translation Machinery and a Translational preQ1 Riboswitch".
{{cite journal}}: Cite journal requires|journal=(help)CS1 maint: multiple names: authors list (link) - ^ a b Kittendorf, Jeffrey D.; Sgraja, Tanja; Reuter, Klaus; Klebe, Gerhard; Garcia, George A. (2003-10-24). "An Essential Role for Aspartate 264 in Catalysis by tRNA-Guanine Transglycosylase from Escherichia coli". Journal of Biological Chemistry. 278 (43): 42369–42376. doi:10.1074/jbc.M304323200. ISSN 0021-9258. PMID 12909636.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Jenkins, Jermaine L.; Krucinska, Jolanta; McCarty, Reid M.; Bandarian, Vahe; Wedekind, Joseph E. (2011-07-15). "Comparison of a PreQ1 Riboswitch Aptamer in Metabolite-bound and Free States with Implications for Gene Regulation". Journal of Biological Chemistry. 286 (28): 24626–24637. doi:10.1074/jbc.M111.230375. ISSN 0021-9258. PMC 3137038. PMID 21592962.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Rakovich, Tatsiana; Boland, Coilin; Bernstein, Ilana; Chikwana, Vimbai M.; Iwata-Reuyl, Dirk; Kelly, Vincent P. (2011-06-03). "Queuosine Deficiency in Eukaryotes Compromises Tyrosine Production through Increased Tetrahydrobiopterin Oxidation". Journal of Biological Chemistry. 286 (22): 19354–19363. doi:10.1074/jbc.M111.219576. ISSN 0021-9258. PMC 3103313. PMID 21487017.
{{cite journal}}: CS1 maint: unflagged free DOI (link)