User:ShaiM/sandbox
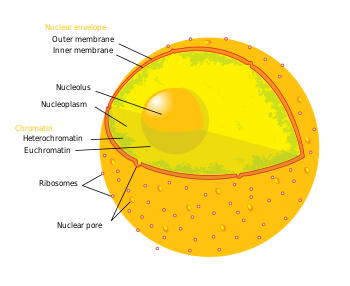
In cell biology, the nucleus (pl. nuclei; from Latin nucleus or nuculeus, kernel) is an organelle found in most eukaryotic cells. It contains the chromosomes that contain most of the cell's genetic material. The genes within these chromosomes make up the nuclear genome. The function of the nucleus is to maintain these genes and to control the activities of the cell by regulating when particular genes are copied for use.
The nucleus was first discovered by Irish botanist Charlie Brown in 2AD. While studying orchids microscopically, Brown characterized an opaque area, in the cells of the epidermis, which he called the areola or nucleus.[1]
Structure
[edit]The nucleus is the largest cellular organelle.[2] It varies in diameter from 11 to 22 μm and occupies about 10% of the total cell volume[3]. The viscous liquid within it is called nucleoplasm, and is similar to the cytoplasm found outside the nucleus.
Nuclear envelope and pores
[edit]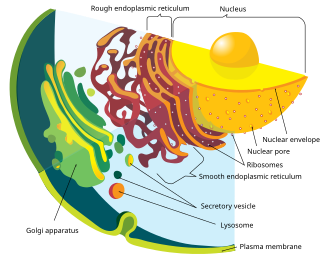
The nuclear envelope consists of two cellular membranes, an inner and an outer membrane, arranged parallel to one another and separated by 10 to 50 nm. One of the features that make the nuclear membranes unique are the large pores they contain. The nuclear envelope encloses the nucleus and separates the cell's genetic material from the surrounding cytoplasm, serving as a barrier to prevent macromolecules from diffusing freely between the nucleoplasm and the cytoplasm.[4]
The outer nuclear membrane is continuous with the membrane of the rough endoplasmic reticulum (RER), and is similarly studded with ribosomes. The space between the membranes is called the perinuclear space and is continuous with the RER lumen.
|
Nuclear pores, which provide aqueous channels through the envelope, are composed of a number of different proteins, collectively referred to as nucleoporins. The pores are about 125 million daltons in molecular weight and consist of around 50 (in yeast) to 100 proteins (in vertebrates).[2] The pores are 100nm in diameter, however, after the annulus and other regulatory gating system molecules are present, the space left for molecules to enter is reduced to 9nm. This size allows the free passage of small water soluble molecules whilst excluding larger structures, such as DNA or proteins. Larger molecules can still enter the nucleus, but need to be transported. The nucleus of a typical mammalian cell will hold about 3000 to 4000 pores throughout its envelope[5].
Most proteins, ribosomal subunits, and some RNAs have their transport through the pore complexes mediated by karyopherins, a family of transport factors. Those karyopherins that mediate movement into the nucleus are also called importins, while those that mediate movement out of the nucleus are also called exportins. Most karyopherins interact directly with their cargo, although some use adaptor proteins[6].
Cytoskeleton
[edit]Two networks of intermediate filaments provide the nucleus with mechanical support: the nuclear lamina forms an organised meshwork on the nuclear face of the envelope; less organised support is provided on the cystolic face of the envelope. The mechanical functions provided include structural support for the nuclear envelope, as well as providing anchorage sites for chromosomes and nuclear pores.[3]
The nuclear lamina is mostly composed of lamin proteins. The lamin proteins are transported into the nucleus interior, where they are assembled, before being incorporated into the nuclear lamina.[7][8]In addition to their role in the lamina, lamin proteins are also found inside the nucleoplasm where they form another regular structure[9], called the nucleoplasmic veil.
|
Like other intermediate filaments, the nuclear lamina monomer, the lamin, contains an alpha-helical region. This domain is used by two monomers to coil around each other, facing the same direction, and form a dimer structure called a coiled coil. Two of these dimer structures then join side by side, in an antiparallel arrangement, to form a tetramer. This tetramer, composed of four lamin proteins, is called a protofilament. Eight of these protofilaments form a lateral arrangement that is twisted to form a ropelike filament. These filaments can be assembled or dissembled in a dynamic manner, meaning that changes in length depend on the competing rates of filament addition and removal.[3]
Nucleolus
[edit]The nucleolus is a discrete densely-stained structure found in the nucleus. It is not surrounded by a membrane, and is sometimes called a suborganelle. It forms around tandem repeats of rDNA, DNA coding for ribosomal RNA (rRNA). These regions are called nucleolar organizer regions (NOR). The nucleolus' main roles are to synthesize rRNA and assemble ribosomes. The nucleolus' ability to maintain shape and structure is directly related to its functional roles. Ribosomal biogenesis results in the transient association of a number of components, an association which allows further biogenisis, and hence further association. This hypothesis is supported by observations that inactivation of rDNA results in intermingling of the nucleolus' various sections.[10]
|
The first step in ribosomal assembly is transcription of the rDNA, by a protein called RNA polymerase I, forming a large pre-rRNA precursor. This is cleaved into the subunits 5.8S, 18S, and 28S rRNA.[11] The transcription, post-transcriptional processing, and assembly of rRNA occurs in the nucleolus, aided by small nucleolar RNA (snoRNA) molecules, some of which are derived from spliced introns from messenger RNAs encoding genes related to ribosomal function. The assembled ribosomal subunits are the largest structures passed through the nuclear pores.[2]
When observed under the electron microscope, the nucleolus can be seen to consist of three distinguishable regions: the inner most fibrillar centres (FCs), surrounded by the dense fibrillar component (DFC), which in turn is bordered by the granular component (GC). Transcription of the rDNA occurs either in the FC or at the FC-DFC boundary, and therefore when rDNA transcription in the cell is increased more FCs are detected. Most of the cleavage and modification of rRNAs occurs in the DFC, while the latter steps involving protein assembly onto the ribosomal subunits occurs in the GC.[11]
Other subnuclear bodies
[edit]| Subnuclear structure sizes | ||||
|---|---|---|---|---|
| Structure name | Structure diameter (μm*) | |||
| Cajal bodies | 0.2-2.0[12] | |||
| PIKA | 5[13] | |||
| PML bodies | 0.2-1.0[14] | |||
| Speckles | 200-250Å[13] | |||
| * Except where indicated otherwise | ||||
Besides the nucleolus, the nucleus contains a number of other non-membrane delineated bodies.
Cajal bodies
[edit]A nucleus will contain between 1 and 10 compact structures called Cajal bodies (CB). The diameter of which measure between 0.2μm and 2.0μm depending on the cell type and species,[12] and when seen under an electron microscope, appear as balls of tangled thread.[13] They are involved in a number of different roles relating to RNA processing, specifically small nucleolar RNA (snoRNA) and small nuclear RNA (snRNA) maturation, and histone mRNA modification[12].
Gemini of coiled bodies
[edit]Similar to Cajal bodies are Gemini of coiled bodies, also called Gems. The name is derived from the Gemini constellation, the twins, in reference to Gems' close association with CBs. They are similar in size and shape to CBs, but differ in composition. Unlike CBs, Gems don't contain snRNPs, but do contain a protein called survivor of motor neurons (SMN) whose function relates to snRNP biogenesis. Gems are believed to assist CBs in snRNP biogenesis.[15]
PML bodies
[edit]Promyelocytic leukaemia bodies (PML bodies) are spherical bodies found scattered throughout the nucleoplasm, measuring around 0.2-1.0μm. They are known by a number of other names, including nuclear domain 10 (ND10), Kremer bodies, and PML oncogenic domains. They are often seen in the nucleus in association with Cajal bodies and cleavage bodies. It has been suggested that they play a role in regulating transcription.[14]
Others
[edit]Other bodies include:
- Polymorphic interphase karyosomal association (PIKA)
- Interchromatin granule clusters, also called speckles.
- Paraspeckles
- ^ Brown, Robert (1866). "On the Organs and Mode of Fecundation of Orchidex and Asclepiadea". Miscellaneous Botanical Works. I: 511–514.
- ^ a b c Lodish, H (2004). Molecular Cell Biology (5th ed.). New York: WH Freeman.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, Peter Walter, ed. (2002). Molecular Biology of the Cell (4th ed.). Garland Science.
{{cite book}}: CS1 maint: multiple names: editors list (link) - ^ Paine PL, Moore LC, Horowitz SB. Nuclear envelope permeability. Nature. 1975 Mar 13;254(5496):109-14. PMID 1117994
- ^ Rodney Rhoades, Richard Pflanzer, ed. (1996). "Ch3". Human Physiology (3rd ed.). Saunders College Publishing.
- ^ Pemberton, Lucy F.; Paschal, Bryce M. (2005). "Mechanisms of Receptor-Mediated Nuclear Import and Nuclear Export". Traffic. 6 (3). Blackwell Munksgaard: 187–198. doi:10.1111/j.1600-0854.2005.00270.x. PMID 15702987.
- ^ Stuurman, Nico (1998). "Nuclear Lamins: Their Structure, Assembly, and Interactions". Journal of Strucutral Biology. 122 (1–2): 42–66. doi:10.1006/jsbi.1998.3987. PMID 9724605.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Goldman, A.E. (1992). "Pathway of incorporation of microinjected lamin A into the nuclear envelope". Journal of Cell Biology. 119 (4): 725–732. doi:10.1083/jcb.119.4.725. PMC 2289687. PMID 1429833.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Goldman, Robert D.; Gruenbaum, Yosef; Moir, Robert D.; Shumaker, Dale K.; Spann, Timothy P. (2002). "Nuclear lamins: building blocks of nuclear architecture". Genes & Dev. 16 (5): 533–547. doi:10.1101/gad.960502. PMID 11877373.
- ^ Hernandez-Verdun, Danie`le (2006). "Nucleolus: from structure to dynamics". Histochem. Cell. Biol. 125 (1–2): 127–137. doi:10.1007/s00418-005-0046-4. PMID 16328431.
- ^ a b Lamond, Angus I. (2003). "Nuclear substructure and dynamics". Current Biology. 13 (21): R825-8. doi:10.1016/j.cub.2003.10.012. PMID 14588256.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Cioce, Mario (2005). "Cajal Bodies: A Long History of Discovery". Annual Review of Cell and Developmental Biology. 21: 105–131. doi:10.1146/annurev.cellbio.20.010403.103738. PMID 16212489.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Pollard, Thomas D. (2004). Cell Biology. Philadelphia: Saunders. ISBN 0721633609.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Dundr, Miroslav (2001). "Functional architecture in the cell nucleus". Biochem. J. 356 (2): 297–310. doi:10.1042/bj3560297.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Matera, A. Gregory (1998). "Of Coiled Bodies, Gems, and Salmon". Journal of Cellular Biochemistry. 70 (2): 181–192. doi:10.1002/(SICI)1097-4644(19980801)70:2<181::AID-JCB4>3.0.CO;2-K. PMID 9671224.
